Data release: OPT imaging of the craniofacial morphology of control and Shroom3 null embryos
Published 15 September 2025
Through integrating Kids First whole genome sequencing with eQTL analyses and supporting 3D imaging data from Shroom3 mouse models, Timothy Cox’s lab has identified SHROOM3 as a bona fide craniofacial microsomia gene. The mouse embryo imaging data from this study is now openly available through FaceBase.
FaceBase Dataset:
Optical Projection Tomography imaging of the craniofacial morphology of control and Shroom3 null embryos
Principal Investigator: Timothy Cox (University of Missouri-Kansas City)
DOI: https://doi.org/10.25550/7E-Y876
Related Publication:
Zhu H, et al. Common cis-regulatory variation modifies the penetrance of pathogenic SHROOM3 variants in craniofacial microsomia. Genome Res. 2025 May 2;35(5):1065-1079. doi: 10.1101/gr.280047.124. PMID: 40234029; PMCID: PMC12047249.
Description:
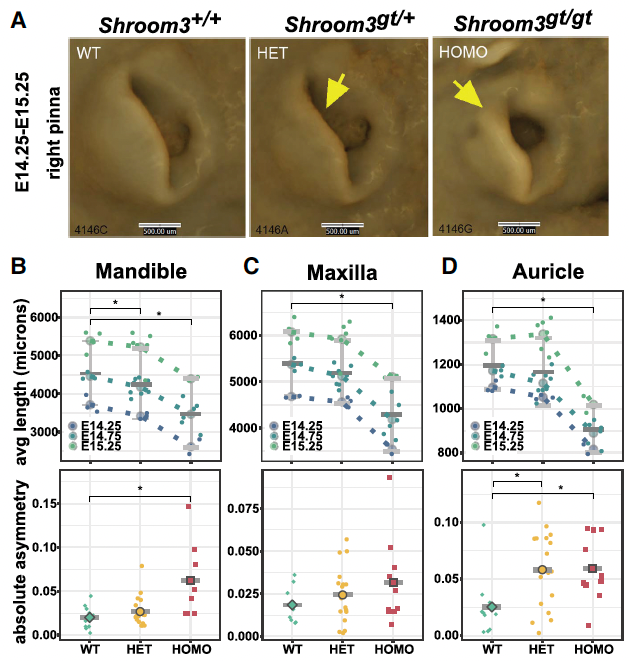
This study followed up on a prior GWAS conducted a large cohort of Chinese patients with craniofacial microsomia (CFM) in which multiple associated genomic loci were identified. Using whole genome sequencing data from 2009 patients with CFM, including data from the Gabriella Miller Kids First program, and 2625 population controls, we analyzed candidate CFM genes within these GWAS peaks and found a significant enrichment of rare variants in one candidate gene, SHROOM3. Subsequent analysis of public expression QTL (eQTL) data revealed numerous eQTLs within the SHROOM3 GWAS peak, prompting us to consider whether eQTLs in combination with the rare variants might determine the presentation of CFM. Allelic phasing identified specific haplotypes in CFM patients, but not in controls, that contained multiple eQTLs in cis with the rare SHROOM3 coding variants. Subsequent luciferase assays confirmed the functional impact of many of these eQTL SNPs. Our findings support the notion that the penetrance of rare variants, that might otherwise be ignored in bioinformatic pipelines because they are seen in unaffected individuals, is influenced by combinations of in cis eQTLs - that is, the expression level of the mutant heterozygous allele may determine the penetrance. Under this premise, we conducted 3D imaging of embryos from two separate Shroom3 mutant lines of mice and performed a morphometric analysis of facial and auricular tissues of wild-type as well as Shroom3 heterozygous and homozygous embryos. These analyses in both models showed Shroom3 gene dosage-dependent effects on both external ear size and maxillary and mandibular growth and, importantly facial asymmetry, providing validation of SHROOM3 as a bona fide CFM gene.
All new sequencing data has been submitted to the National Genomics Data Center (https://ngdc.cncb.ac.cn/gsa-human) under accession numbers HRA005132, HRA004333, HRA003925, and HRA003924. Raw, reconstructed, and rendered optical projection tomography (OPT) data for all mouse embryos of the Shroom3gt strain used for the morphometric analyses is available now on FaceBase (https://doi.org/10.25550/7E-Y876). MicroCT datasets of the Shroom3 null allele strain are available through the IMPC website with IDs 157760 (wildtype) and 612303 (homozygous) (https://www.mousephenotype.org/embryoviewer/?mgi=MGI%3A1351655).
