News & Events
New release: MicroCT analysis of ameloblast specific Smad4 conditional knockout mice and Ambn-IRESCre mouse models (11 February 2026)
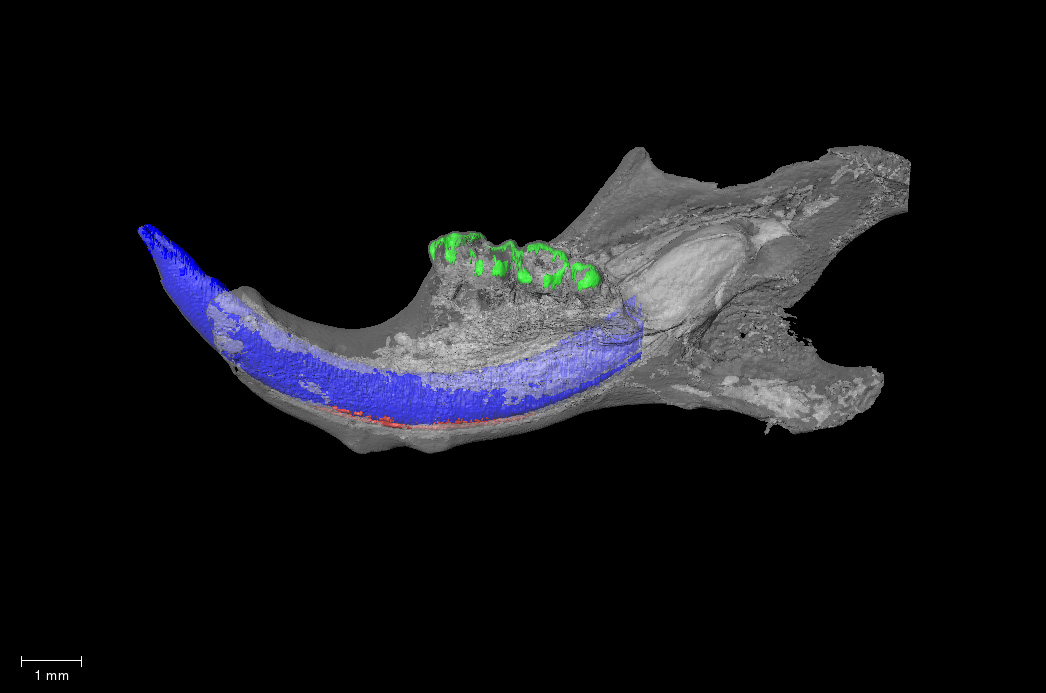 Mutant sample from the dataset MicroCT analysis of ameloblast specific Smad4 conditional knockout mice
Mutant sample from the dataset MicroCT analysis of ameloblast specific Smad4 conditional knockout mice
Description:
Michael Paine’s lab (University of Southern California) developed a novel ameloblastin driven Cre-recombinase mouse line (Ambn-IRESCre MMRRC Strain #067446-JAX; RRID:MMRRC_067446-JAX) with Cre expression limited to enamel forming cells called ameloblasts. This is a significant advance from the current Cre mouse lines used in conditional gene silencing to study enamel formation. We used this Ambn-IRESCre line to knockdown the transcription factor Smad4 specifically in ameloblasts and achieved an extreme pathological phenotype limited to tooth enamel with no effects on the physiological functions of Smad4. These data highlight the significance of the Ambn-IRESCre mouse model for future studies defining gene functions during enamel formation.
Related Resource:
This data is part of EnamelBase, a community resource providing validated genetic mouse tools and multi-scale data to study gene expression, structure, and mechanical properties of developing and mature tooth enamel.
FaceBase Datasets:
Michael Lansdell Paine, Rucha Arun Bapat, Alexis Bauer, David Evans, Yanbin Ji, Marziyeh Aghazadeh. MicroCT analysis of ameloblast specific _Smad4_ conditional knockout mice. FaceBase Consortium https://doi.org/10.25550/2W-YN1C (2026).
Rucha Arun Bapat, Alexis Bauer, David Evans, Marziyeh Aghazadeh, Yanbin Ji, Michael Lansdell Paine. MicroCT analysis of Ambn-IRESCre mouse models. FaceBase Consortium https://doi.org/10.25550/7X-HZDA (2026).
Registration and Call for Abstracts Now Open: FaceBase Community Forum 2026 (04 February 2026)
We’re pleased to announce that registration and the Call for Abstracts are now open for the FaceBase Community Forum 2026!
Monday–Tuesday, May 4–5, 2026
Arlington, VA (USC/ISI East Office)
Theme
Advancing Reuse, Integration, and AI-Ready Research Across Craniofacial and Related Health Domains to Enhance Whole Person Health
This year’s Forum will bring together researchers, clinicians, data scientists, trainees, and data contributors to explore how shared, well-curated data and modern data science approaches can accelerate discovery across dental, oral, craniofacial, inner ear, and related health domains.
What to expect
- Keynote talks and NIH program updates
- Sessions on data federation, collaboration, and AI-ready data resources
- Panels on clinical integration and the FaceBase EMR pilot project
- A poster session highlighting community-driven work and data reuse examples
Call for Abstracts We invite submissions describing completed studies, ongoing work, methods, tools, or emerging ideas related to data reuse, integration, and AI-ready research. Abstracts will be used to organize the poster session, and a subset may be selected for invited talks.
Register and submit here:
We hope you’ll join us in Arlington this May and help shape the conversation around data reuse and AI-ready biomedical research.
New release: Gene expression patterns of the developing face at single cell resolution (20 January 2026)
The following datasets have been released to FaceBase by Dr. Justin Cotney’s lab of the Children’s Hospital of Philadelphia. A related manuscript has been accepted but not yet released; in the meantime, these data are available for the community.
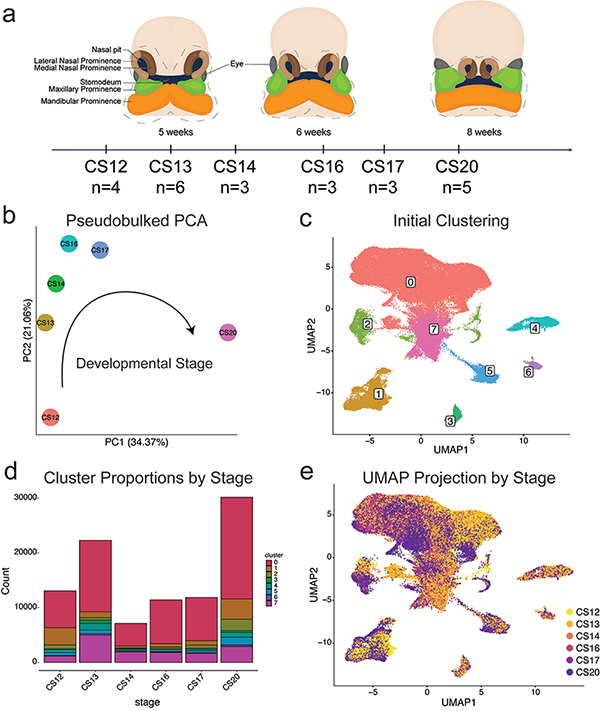
Description:
Justin Cotney’s lab generated a single-nucleus RNA sequencing (snRNA-seq) atlas of critical time points during human and mouse craniofacial development. Their analysis identified multiple subtypes of mesenchyme, epithelium, and cranial neural crest, among other functionally distinct cell types. Through cross-species comparisons, Cotney’s team identified functionally conserved cell types as well as anatomically distinct cell subtypes. Finally, following integration with data from the Gabriella Miller Kids First program, the Cotney lab revealed enrichment of de novo protein-altering variants from orofacial clefting trios in specific cell subtypes. The human and mouse snRNA-seq data from this study is now openly available through FaceBase.
FaceBase Datasets:
Alexandra Manchel, Justin Cotney. Gene expression patterns of the developing human face at single cell resolution (snRNA-seq). FaceBase Consortium https://doi.org/10.25550/8D-2JQ0 (2026).
Alexandra Manchel, Justin Cotney. Gene expression patterns of the developing mouse face at single cell resolution (snRNA-seq). FaceBase Consortium https://doi.org/10.25550/7N-54BY (2026).
Save the Date: 2026 FaceBase Community Forum – May 4–5 in Arlington, VA (11 December 2025)
We’re excited to announce that the 2026 FaceBase Community Forum will take place May 4–5, 2026, at the University of Southern California/Information Sciences Institute East Office in Arlington, VA. This two-day, in-person event will bring together researchers, clinicians, data scientists, contributors, and trainees from across dental, oral, and craniofacial (DOC) science as well as related areas such as the ear, eye, and other craniofacial-associated systems.
This year’s annual meeting will highlight new tools, datasets, and research supported by FaceBase, with expanded discussions on AI-ready biomedical data and how structured metadata, packaging, and standards within the platform enable machine learning, multimodal integration, and new scientific insights.
Sessions will include invited talks, panel discussions, demonstrations, poster presentations, and community-driven conversations with the FaceBase team. It’s an opportunity to learn, collaborate, and help shape the future of the FaceBase resource.
An agenda, registration information, and the call for posters will be available early 2026.
For more details and updates, visit the event page: https://www.facebase.org/community/annual-meeting/2026-facebase-annual/
Materials Now Available: November 2025 FaceBase Bootcamp (10 November 2025)
Thank you to everyone who joined our November 2025 FaceBase Bootcamp for Users and Data Contributors!
Recordings, slides, and other materials from the session are now available on the event page.
The bootcamp covered:
- How to find, filter, and download data from FaceBase
- Requesting controlled-access human datasets
- Preparing and contributing data using FaceBase tools
- Guidance for NIH Data Management and Sharing (DMS) plans
- And more!
Watch individual segments or the full playlist on our YouTube channel, and download the presentation slides directly from the event page.
We’d love your feedback!
If you took the training (either live or later via video) please take our short, anonymous 3-minute survey to help shape future FaceBase trainings.

