Human Genomics Analysis Interface
This project is a genome-wide meta-analyses of cleft lip with or without palate (CL/P), cleft palate alone (CP), and all orofacial clefts (OFCs) across two large, multiethnic studies, GENEVA OFC and POFC, both of which can be explored through our Human GenomicsAnalysis Interface.
For information about access to individual level data via dbGaP, study investigators and funding sources, please see the respective pages of the two original projects, GENEVA OFC and POFC .
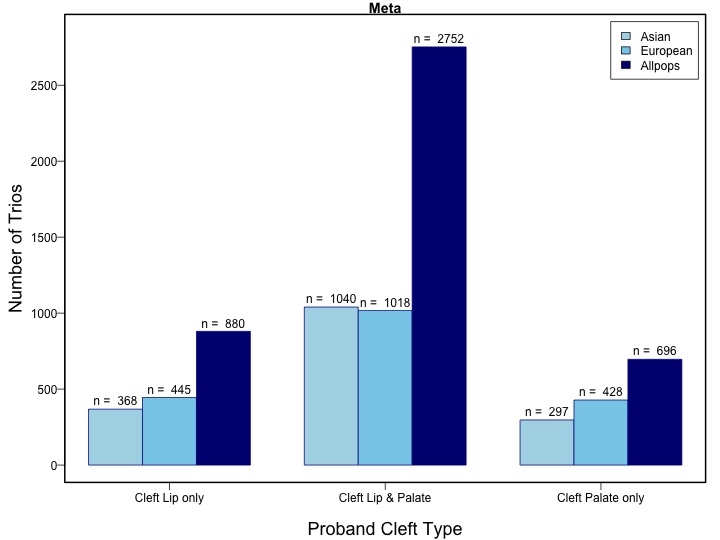
The goal of this study is to identify additional genetic risk variants for OFCs, considering the historical groupings of CL/P and CP, but also exploring the possibility of shared etiology. Therefore, we conducted genome-wide meta-analyses for CL/P, CP, and all OFCs, drawing from the two of the largest CL/P studies published to date and the two published CP studies. In addition to the combined analysis of the two multiethnic study populations, we also conducted population specific meta-analyses in sub-samples of Asian and European ancestry.
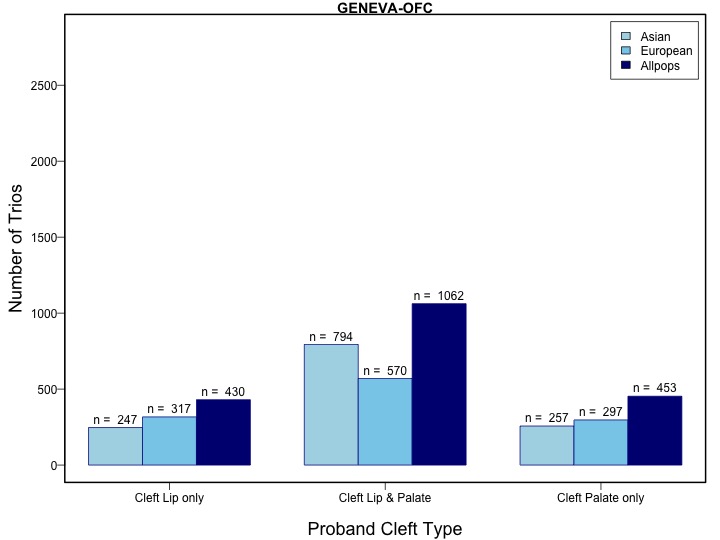
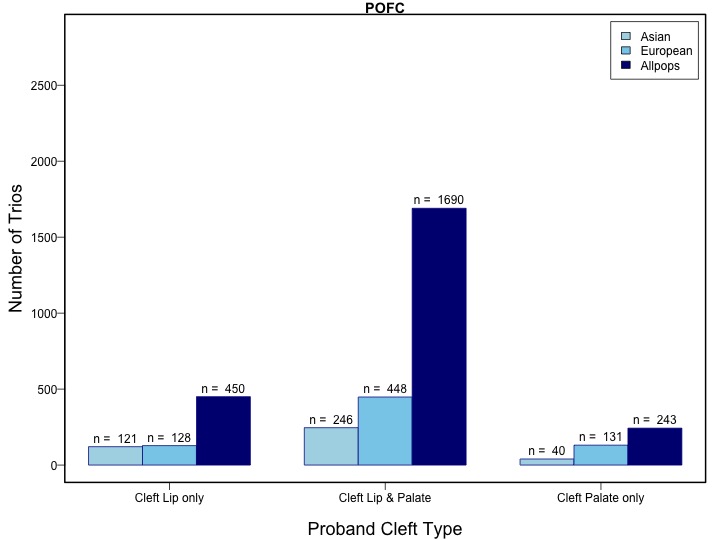
A total of 412 individuals, who were common to both the GENEVA OFC and POFC studies were excluded from the GWASs of the GENEVA OFC study for this analysis prior to meta-analysis.
Methods
For study-specific information such as genotyping and quality control methods, please see the respective pages of the original projects, GENEVA OFC and POFC.
The two case-parent trio subgroups from POFC and GENEVA OFC were analyzed separately using the transmission disequilibrium test (TDT), available in the PLINK software. The resulting effects estimates for the three analysis groups were combined in an inverse variance-weighted fixed-effects meta-analysis using R. Stratified meta-analysis was conducted for Asian and European subpopulations as determined by ancestry principal component analysis (PCA).
A total of 412 individuals were in both the GENEVA OFC and POFC studies so these participants were excluded from the GWASs of the GENEVA OFC study for this analysis prior to meta-analysis.
Meta analysis Manhattan plots were created using the qqman2 package in R.
Explore Project Data
The samples for this study come from four primary ancestry groups: Europeans (from the United States, Denmark, Hungary, Spain, and Turkey), Asians (from China, the Philippines, and India), Central/South Americans (from Puerto Rico, Guatemala, Colombia, and Argentina), and Africans (from Nigeria and Ethiopia). OFC phenotypes include cleft lip with or without cleft palate (CL/P), cleft palate only (CP) and all OFCs combined (All).
All genomic coordinates are mapped to Human GRCh37 - hg19 genome assembly.
Custom Plots
Please select one option from each of the categories. You must select one option from each available category to be able to submit the request.Publications
PubMed list for all publications associated with this project will go here.