Human Genomics Analysis Interface
dbGaP Study Accession: phs000949.v1.p1
Principal Investigator: Seth Weinberg , University of Pittsburgh School of Dental Medicine, Pittsburgh, PA, USA
Funding Source: U01-DE020078, National Institute of Dental and Craniofacial Research, Bethesda, MD, USA
U01-DE020057, National Institute of Dental and Craniofacial Research, Bethesda, MD, USA
R01-DE016148, National Institute of Dental and Craniofacial Research, Bethesda, MD, USA
U01-DE020054, National Institute of Dental and Craniofacial Research, Bethesda, MD, USA
U01-DE024425, National Institute of Dental and Craniofacial Research, Bethesda, MD, USA
K99-DE02560, National Institute of Dental and Craniofacial Research, Bethesda, MD, USA
Contract #HHSN268201200008I, National Institute of Dental and Craniofacial Research, Bethesda, MD, USA
X01-HG007821, National Human Genome Research Institute, Bethesda, MD, USA
Although ample evidence exists that facial appearance and structure are highly heritable, there is a dearth of information regarding how variation in specific genes relates to the diversity of facial forms evident in our species. With the advent of affordable, non-invasive 3D surface imaging technology, it is now possible to capture detailed quantitative information about the face in a large number of individuals. By coupling state- of-the-art 3D imaging with advances in high-throughput genotyping, an unparalleled opportunity exists to map the genetic determinants of normal facial variation. The project contains the results of GWAS performed on 3D facial measurements from two cohorts of US Caucasians collected as part of the FaceBase consortium. These results were published in PLoS Genetics by Shaffer et al. (2016).
Methods
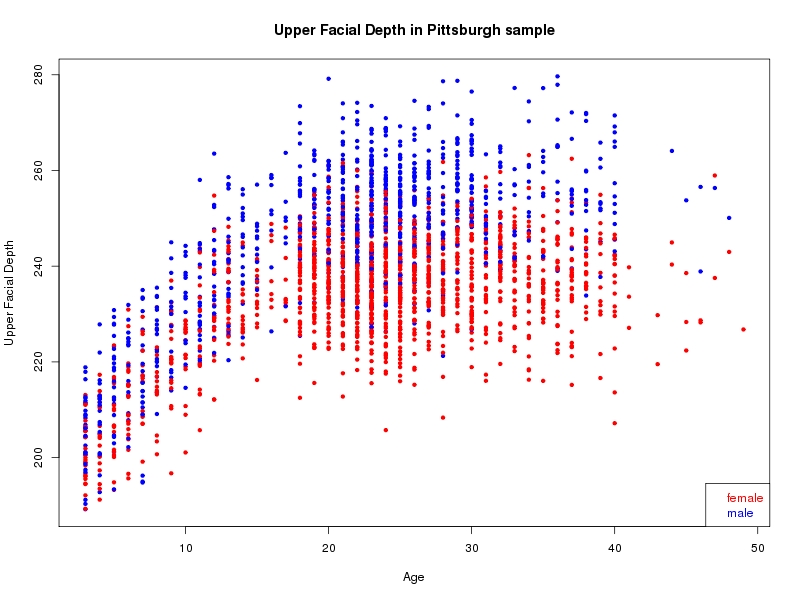
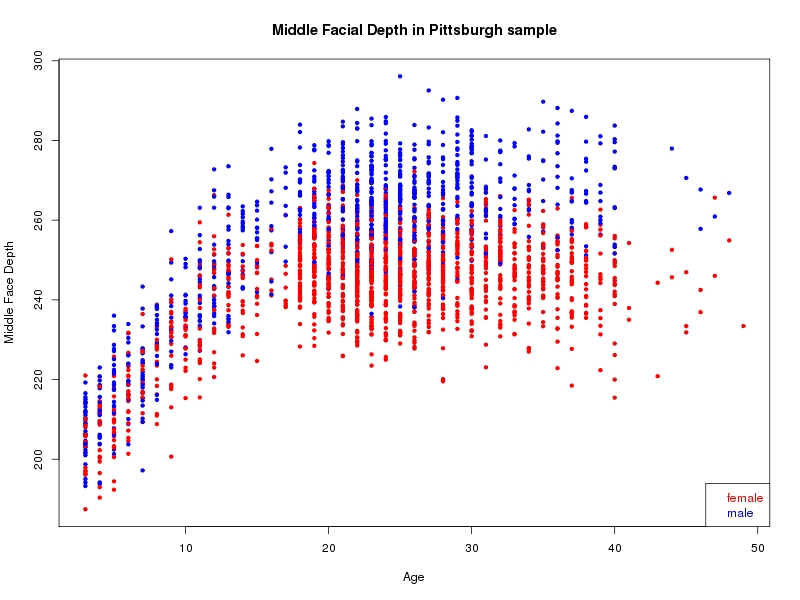
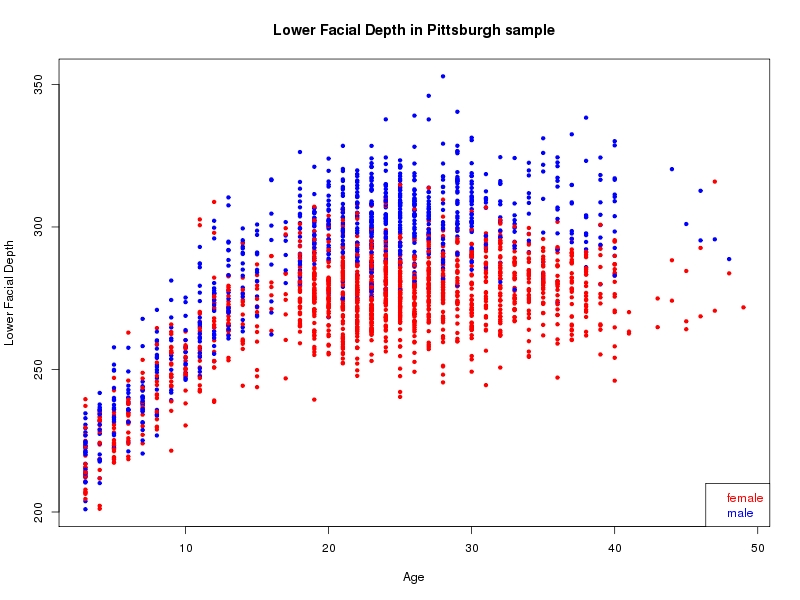
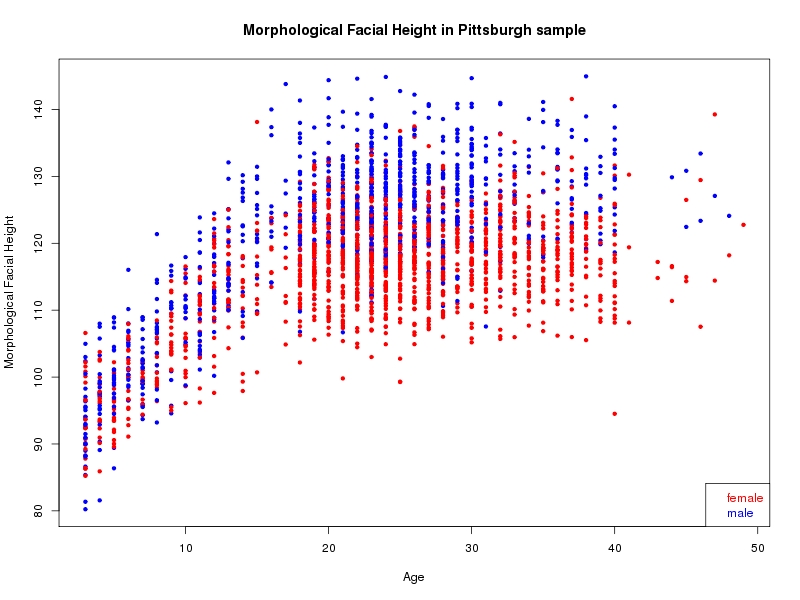
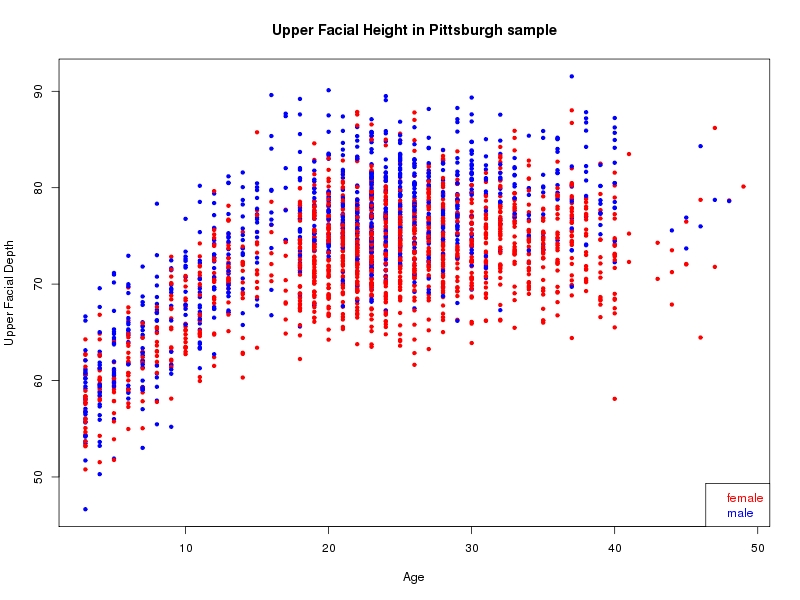
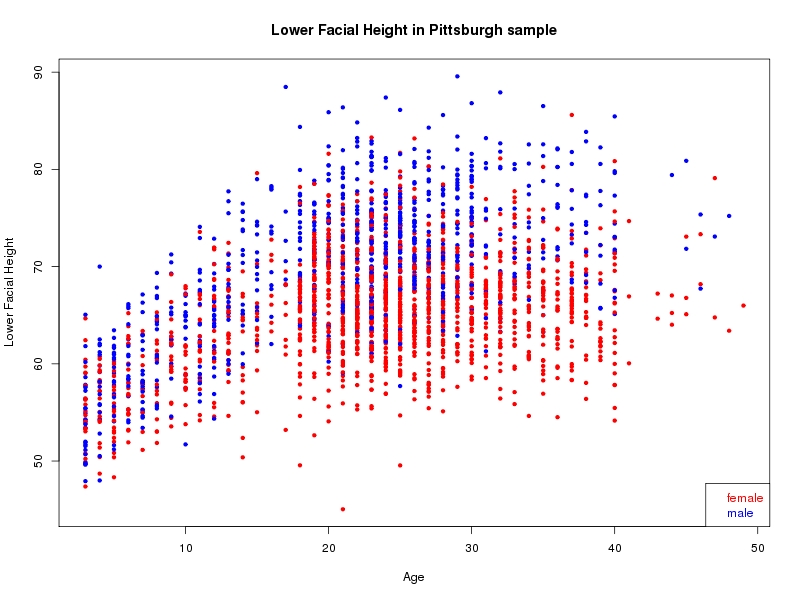
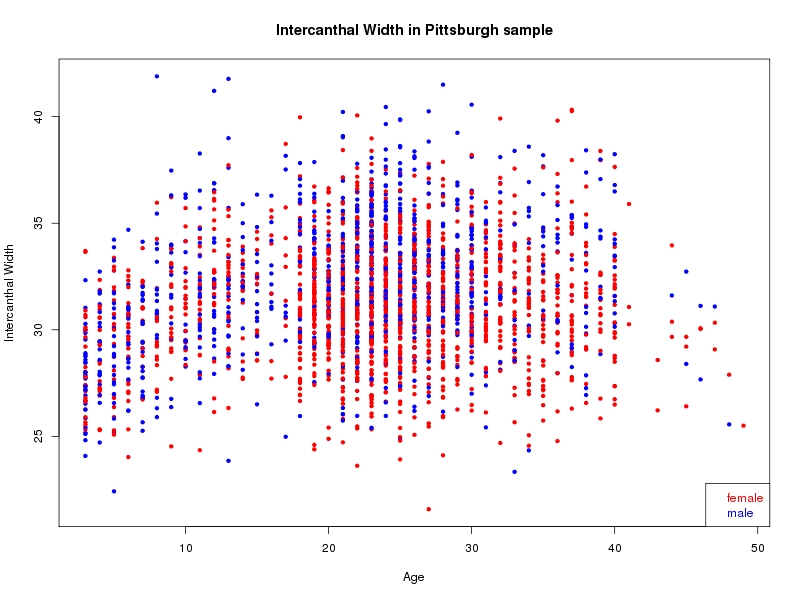
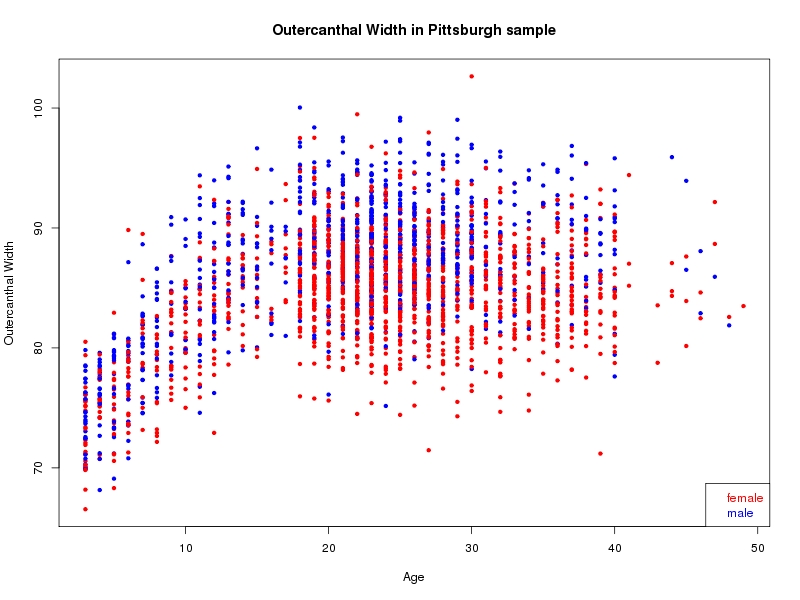
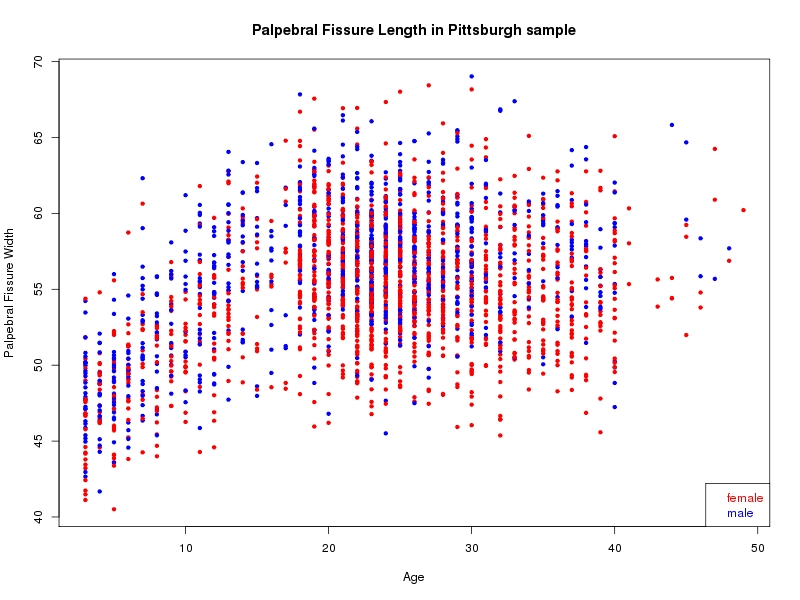
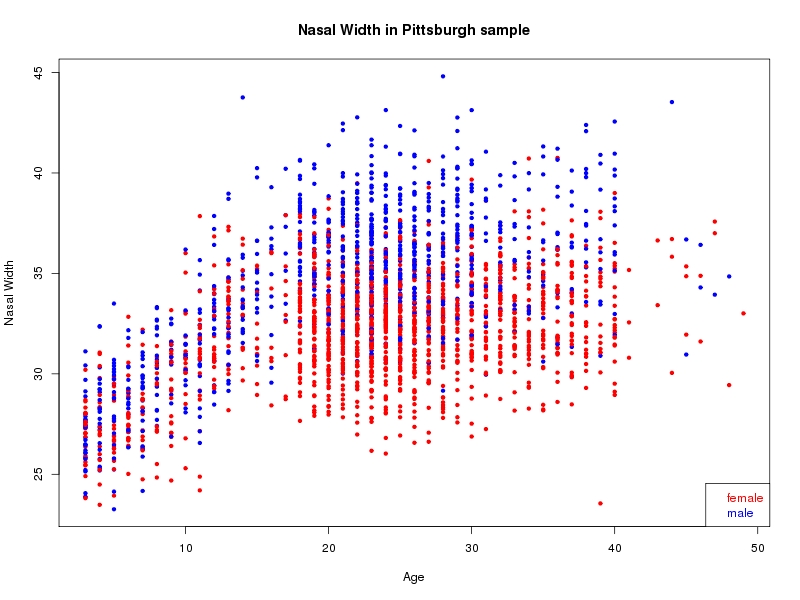
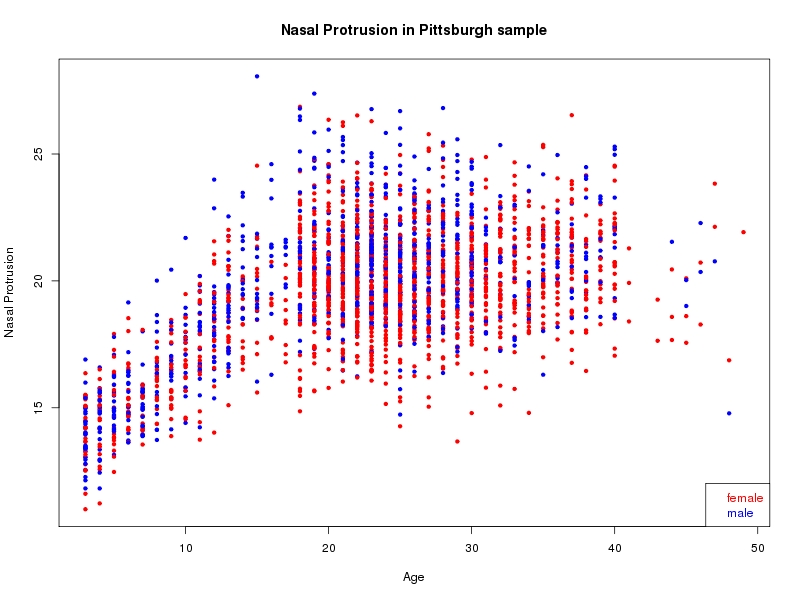
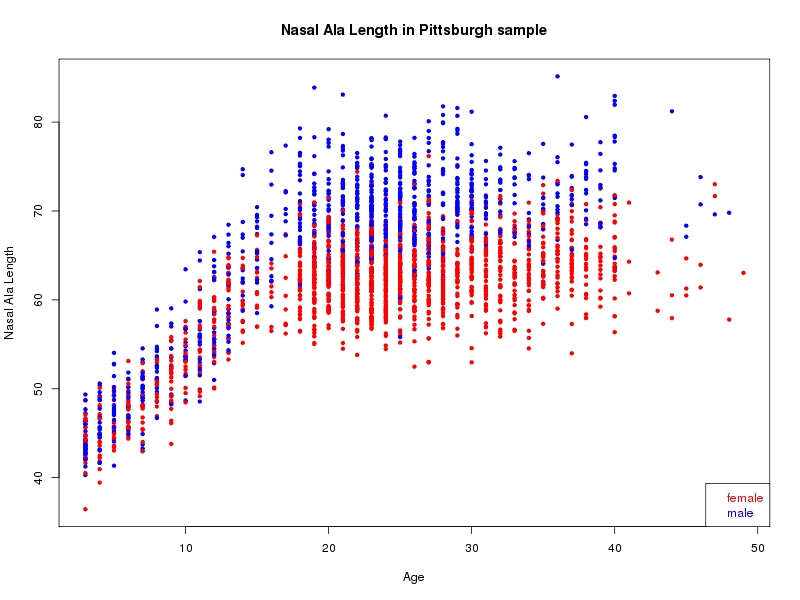
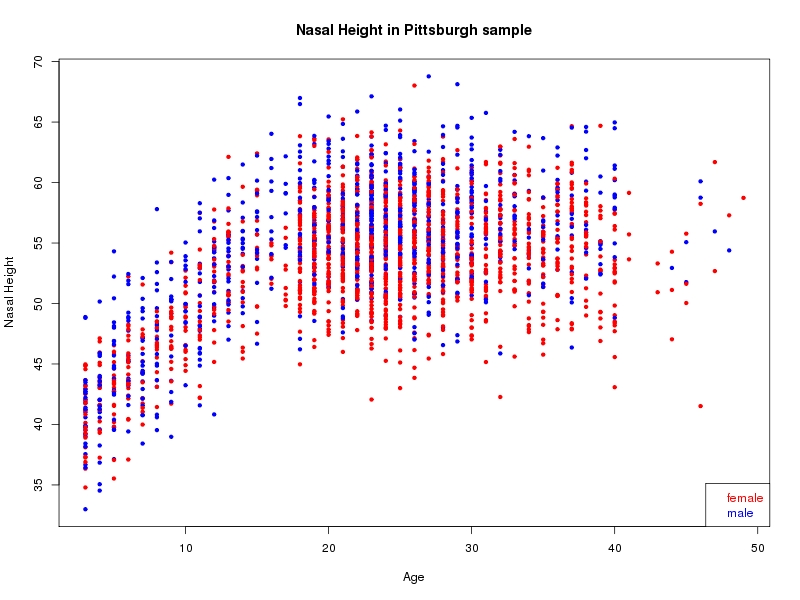
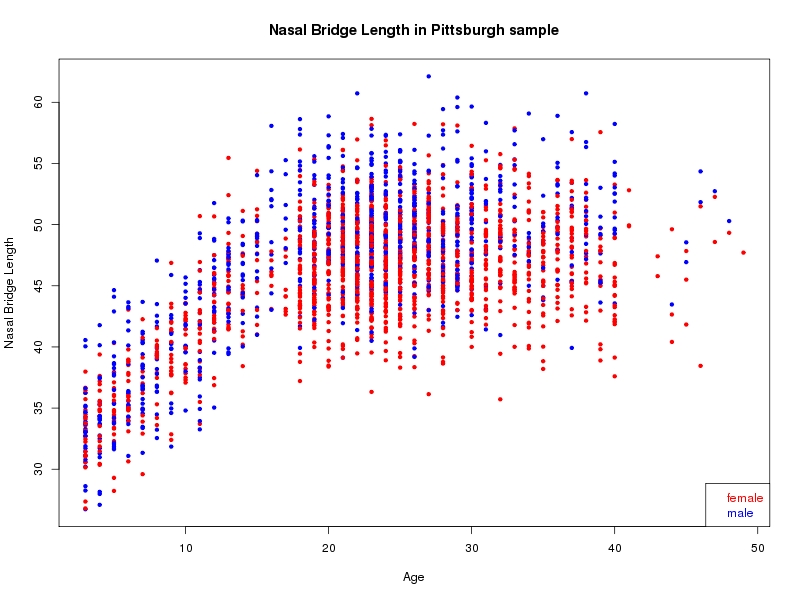
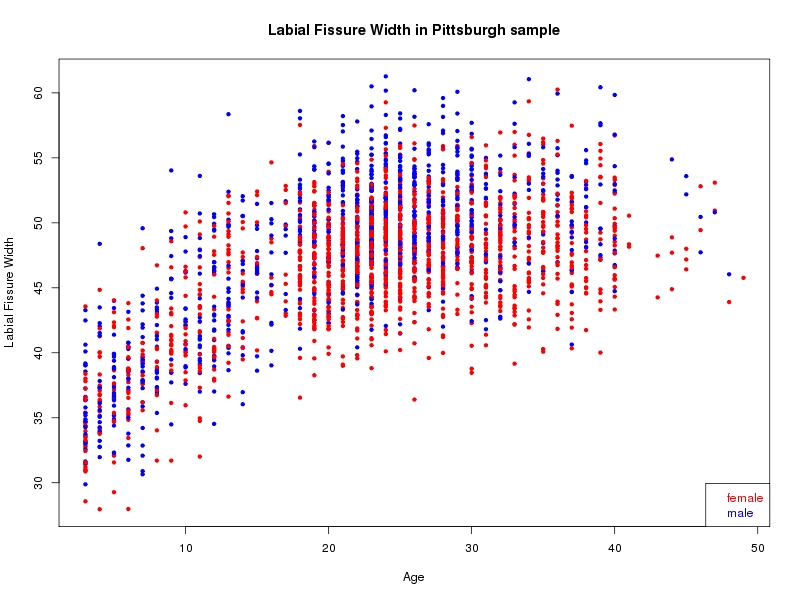
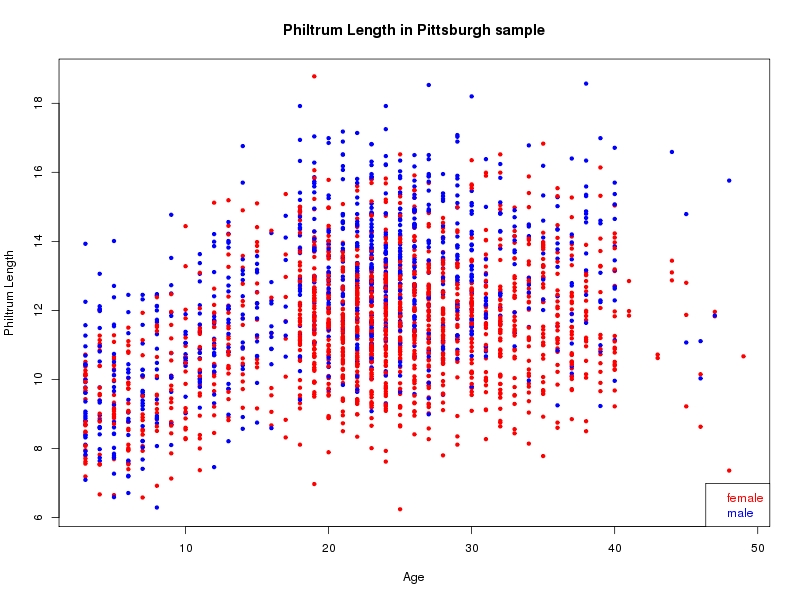
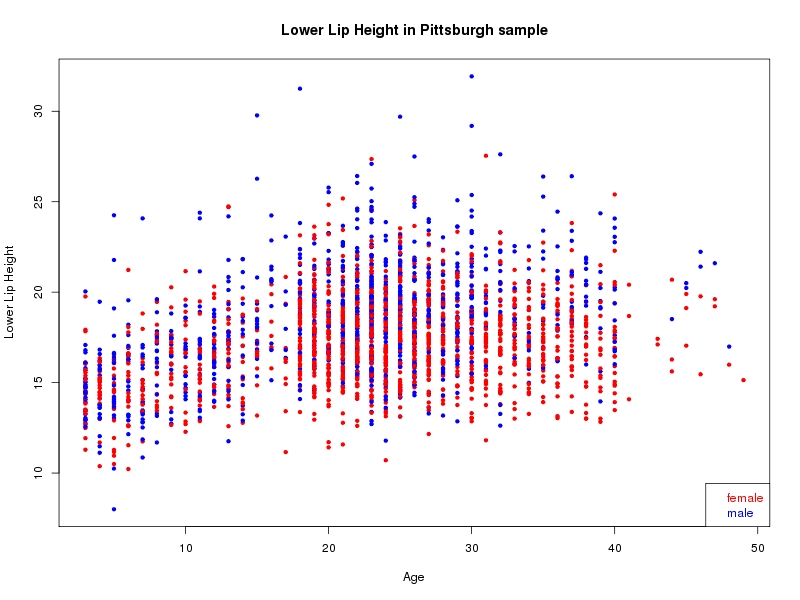
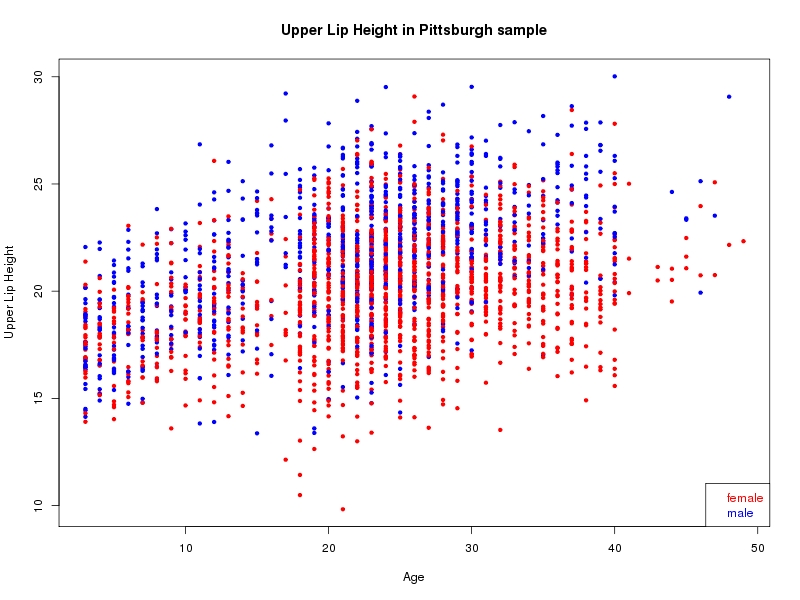
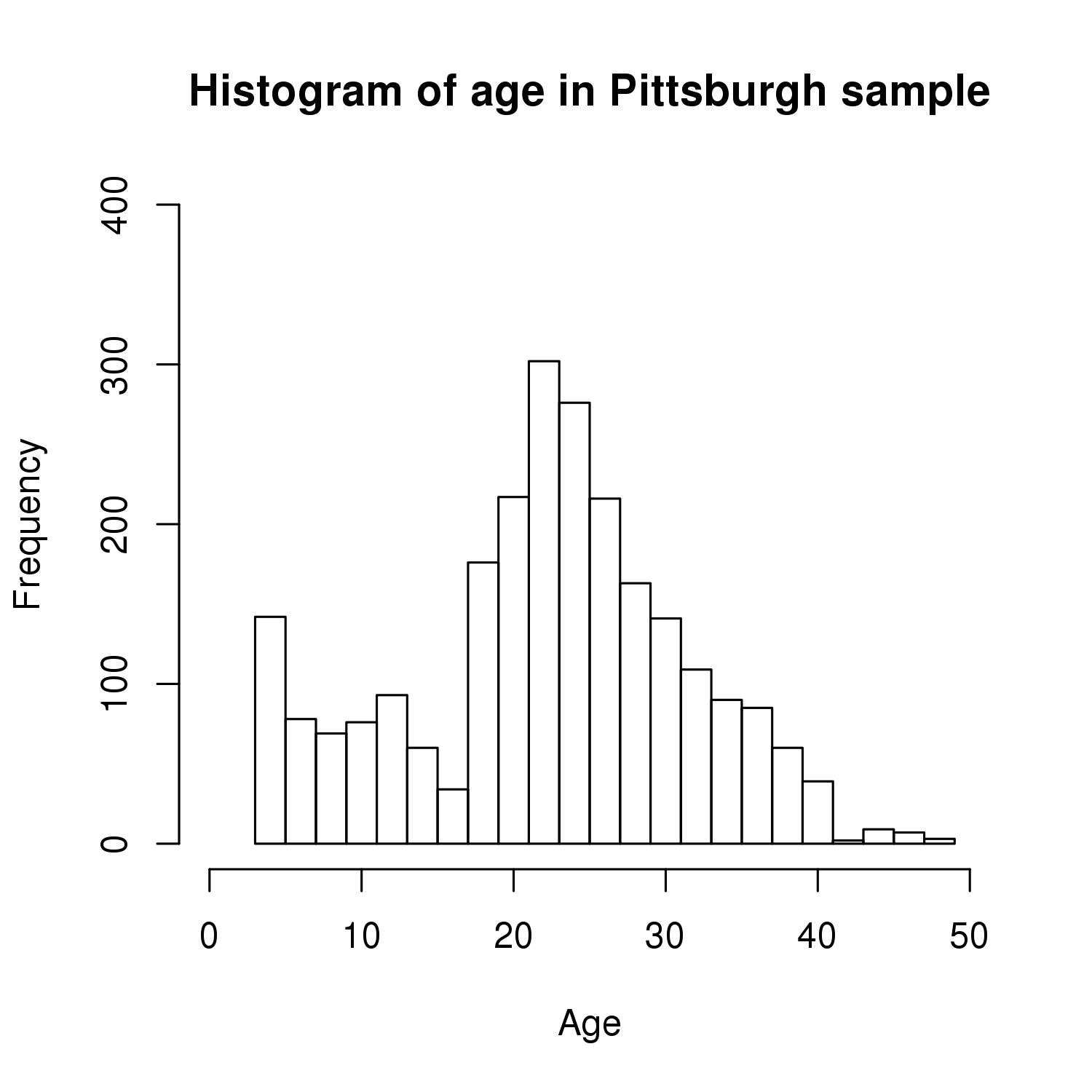
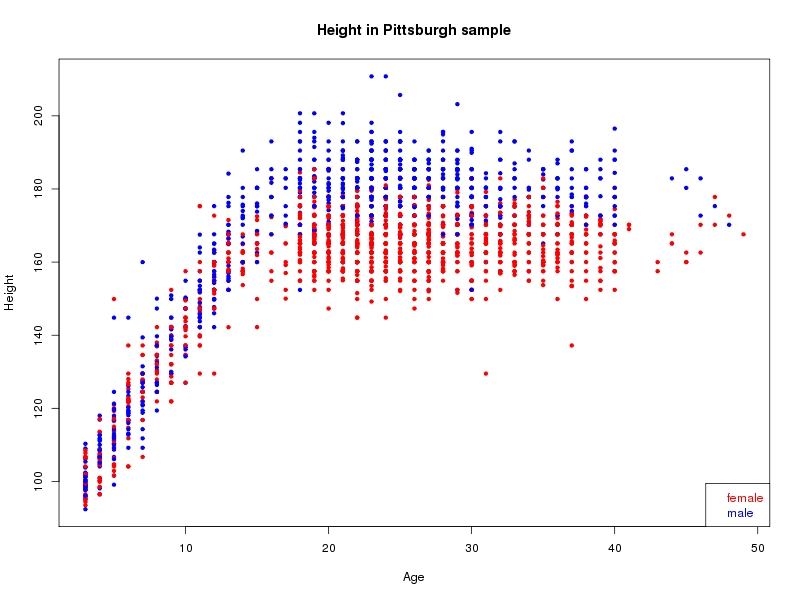
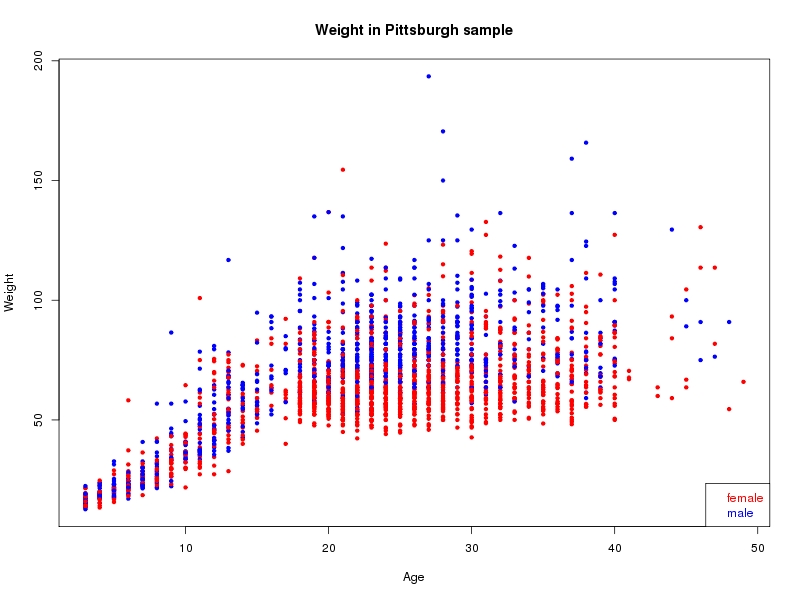
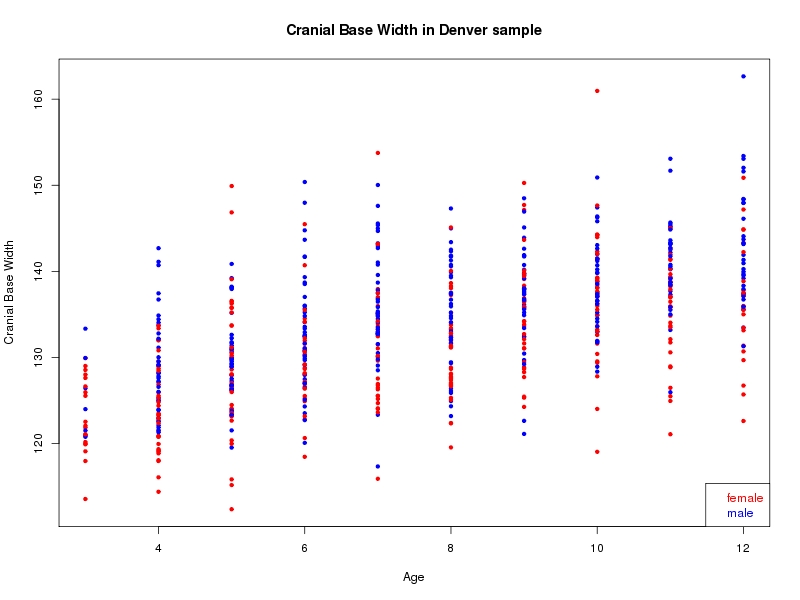
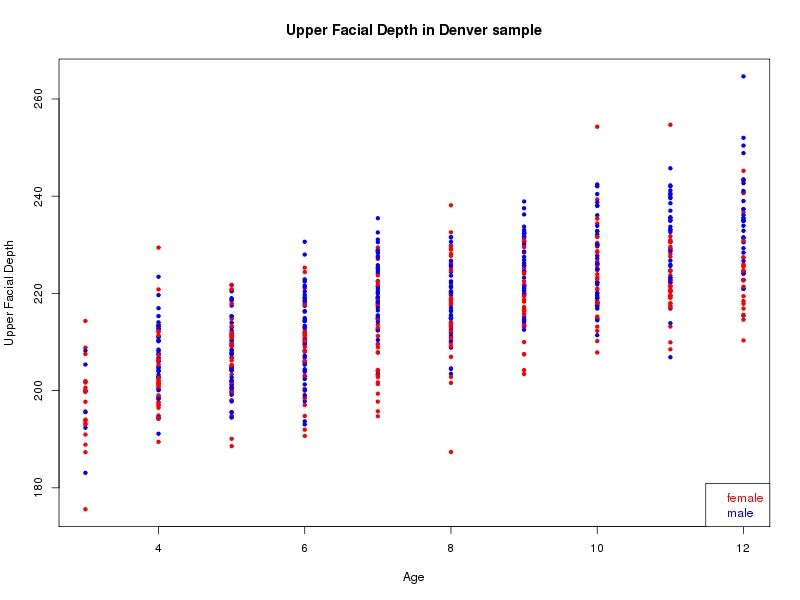
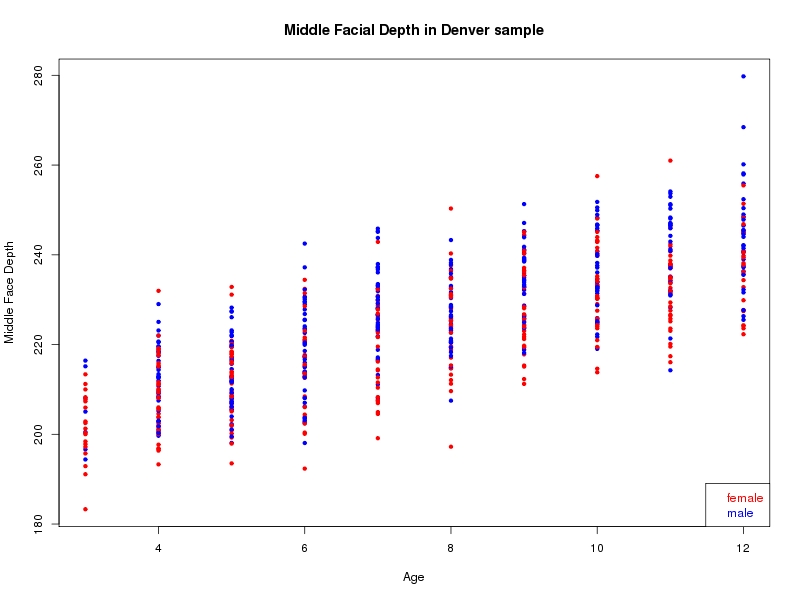
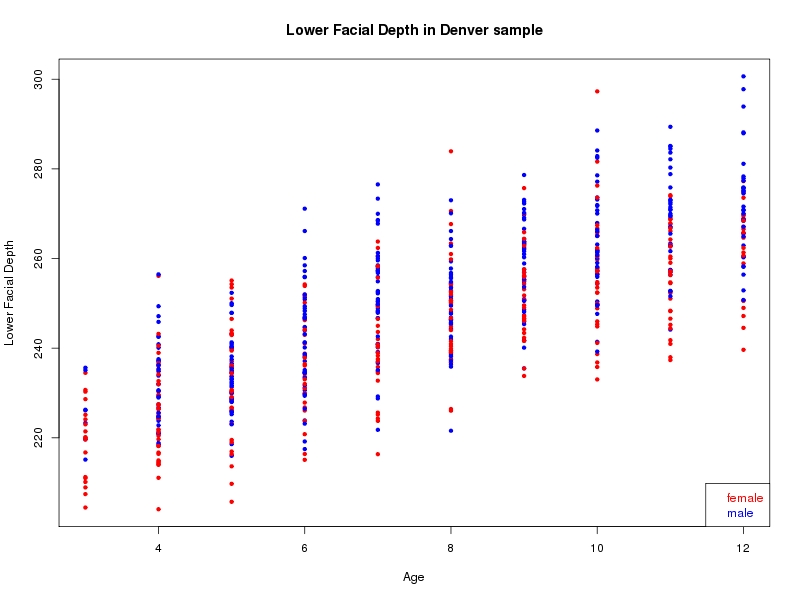
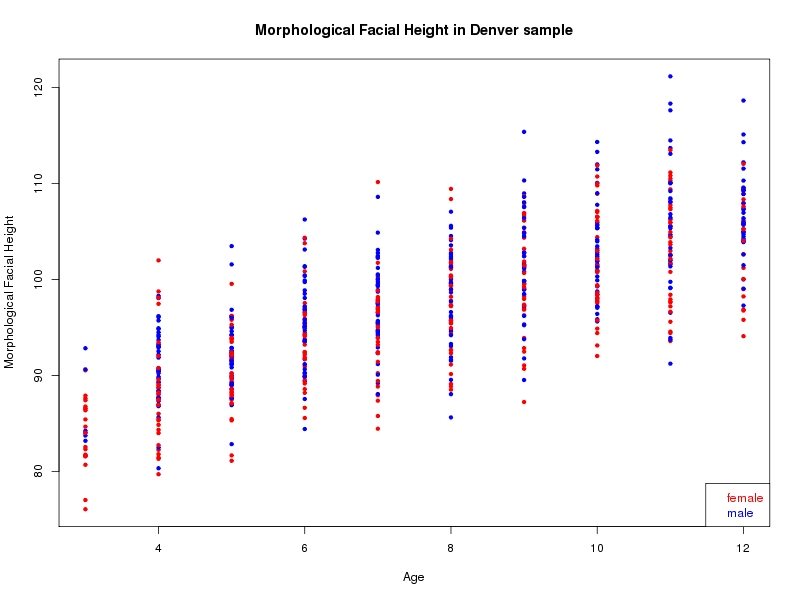
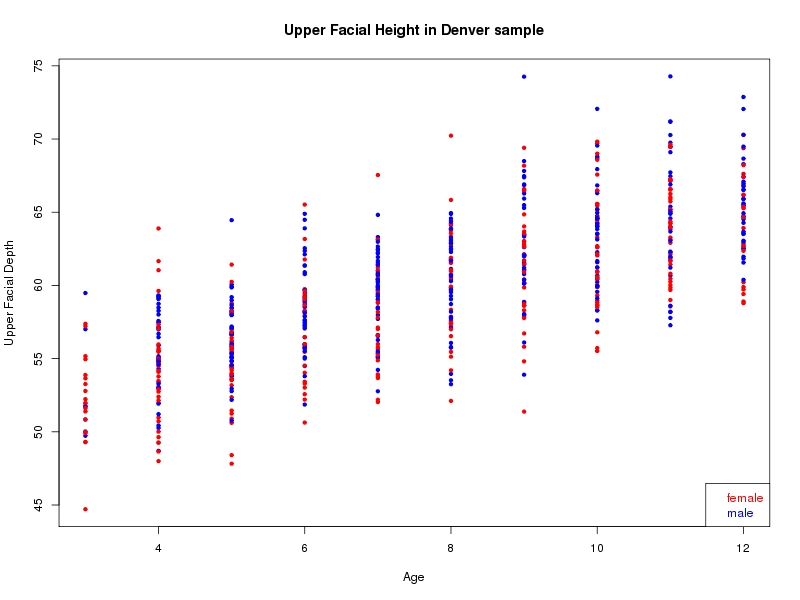
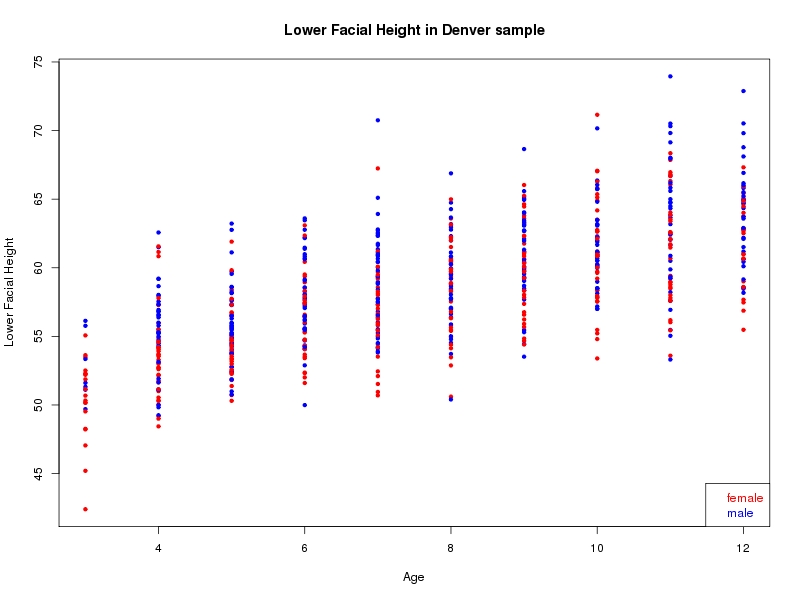
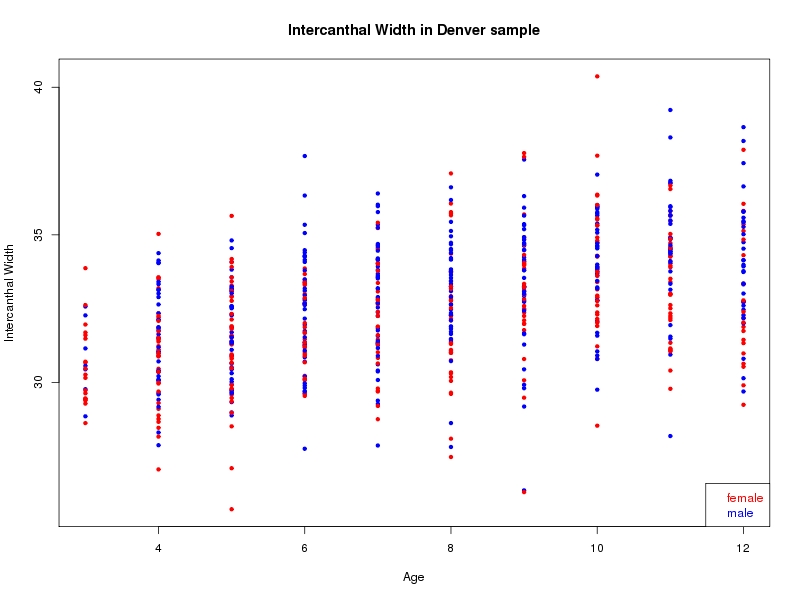
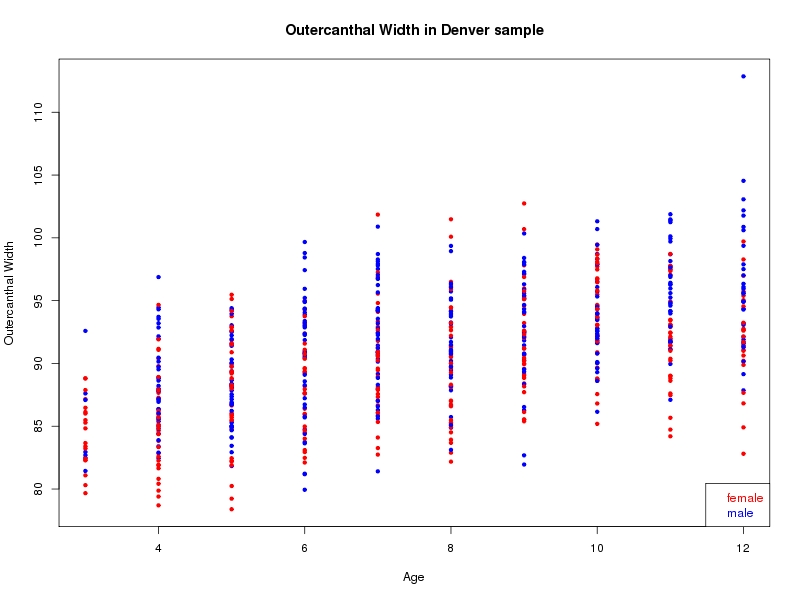
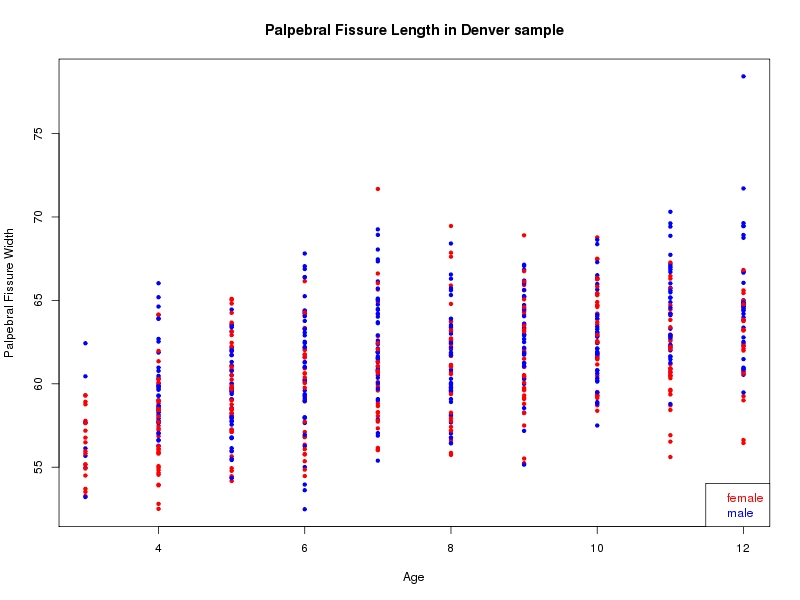
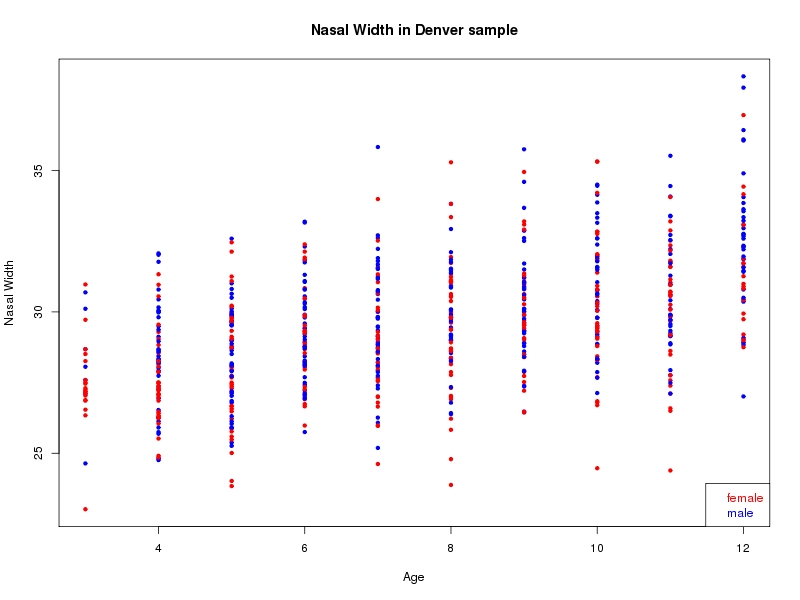
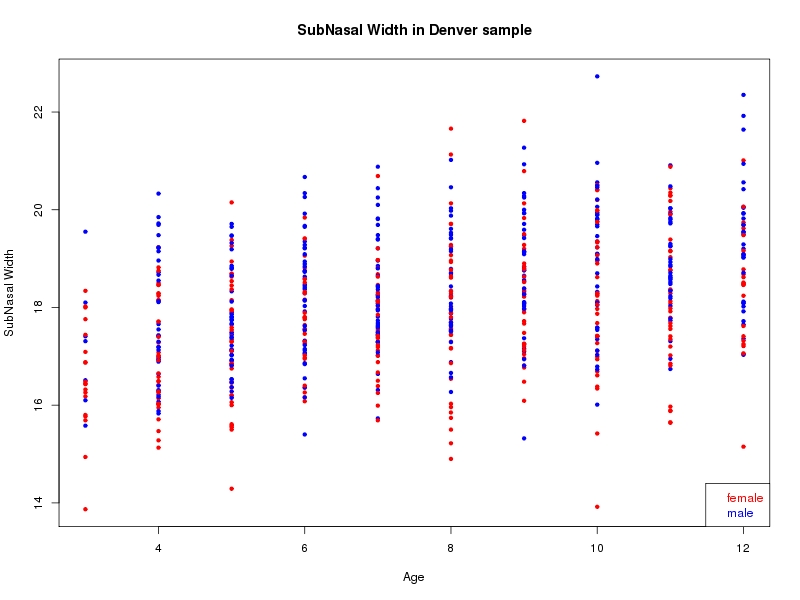
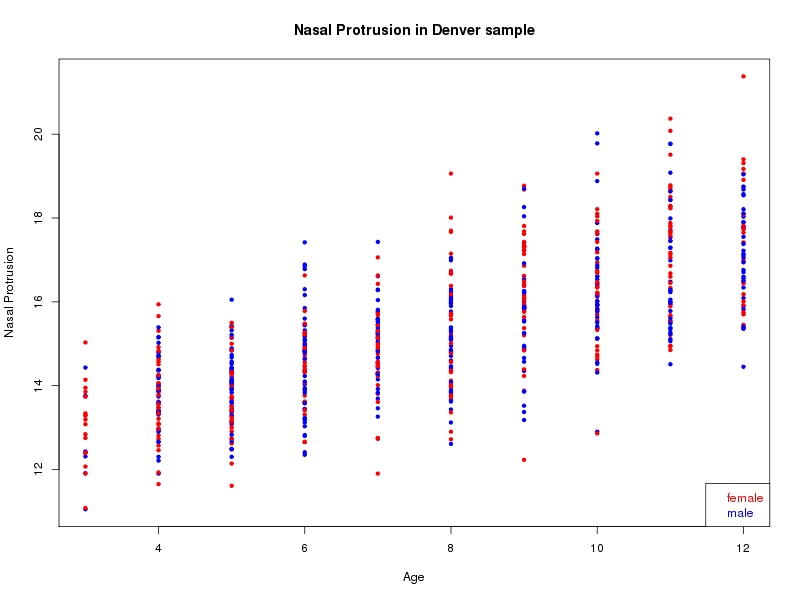
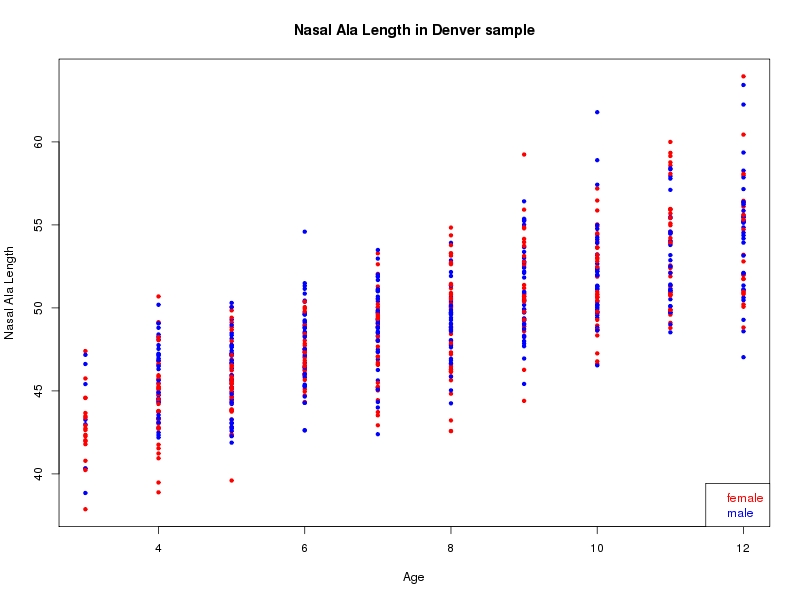
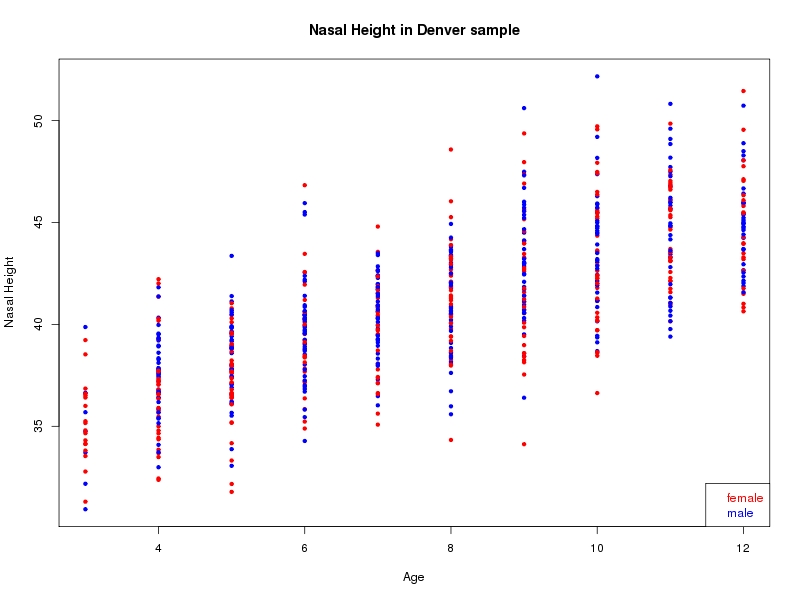
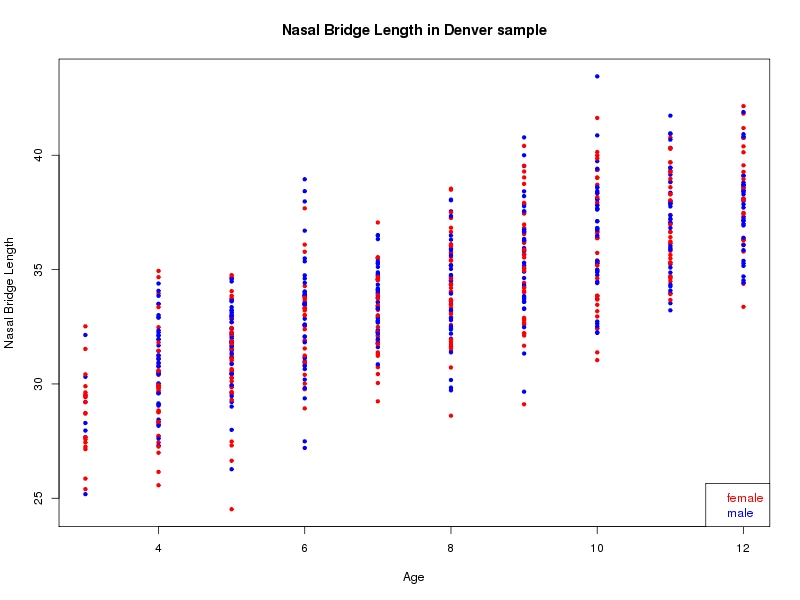
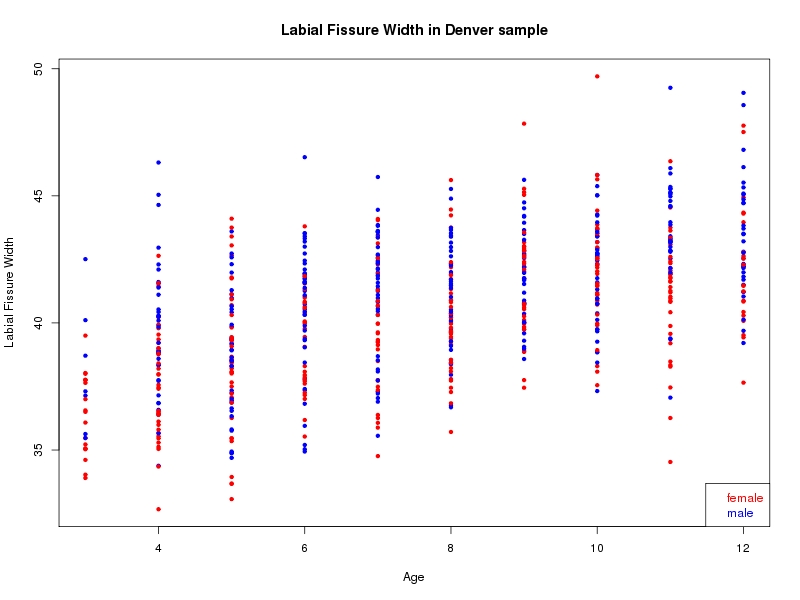
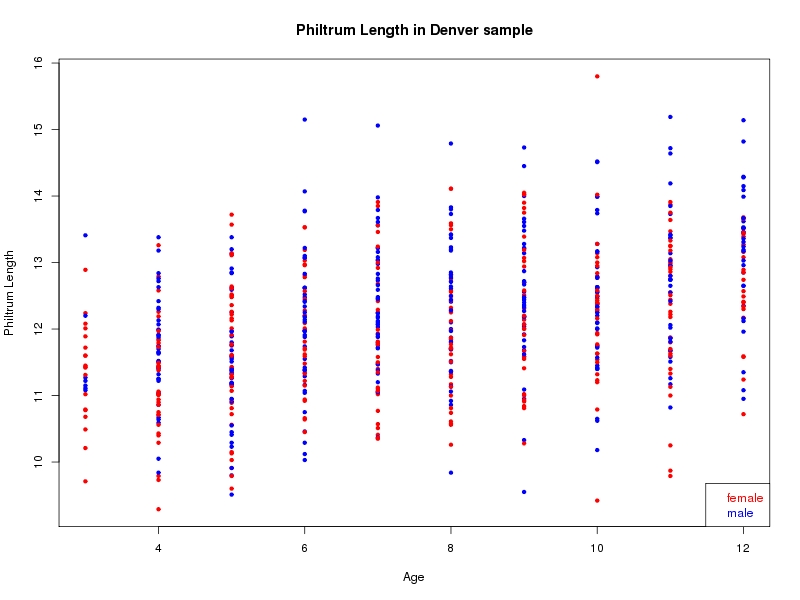
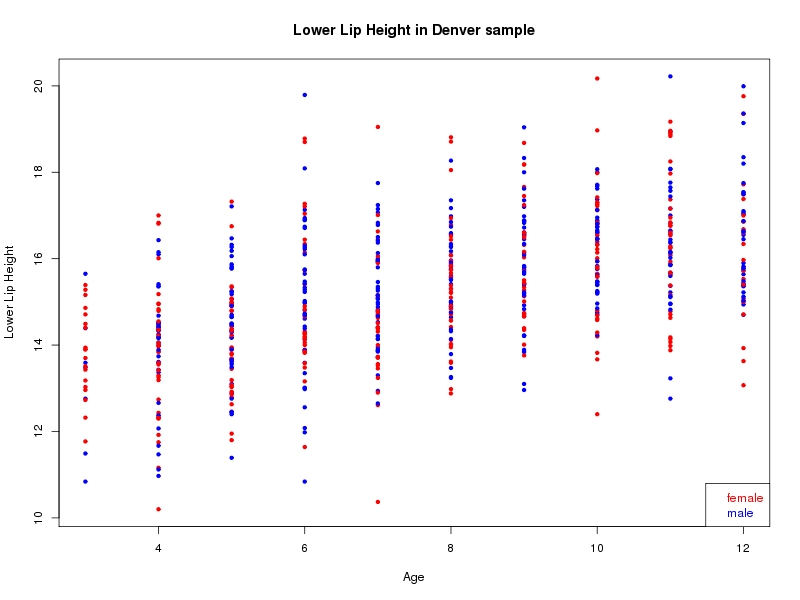
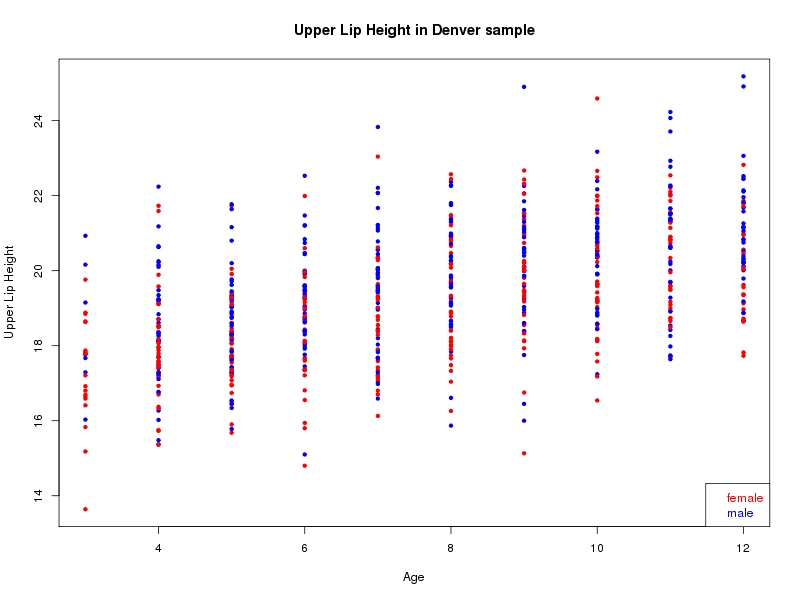
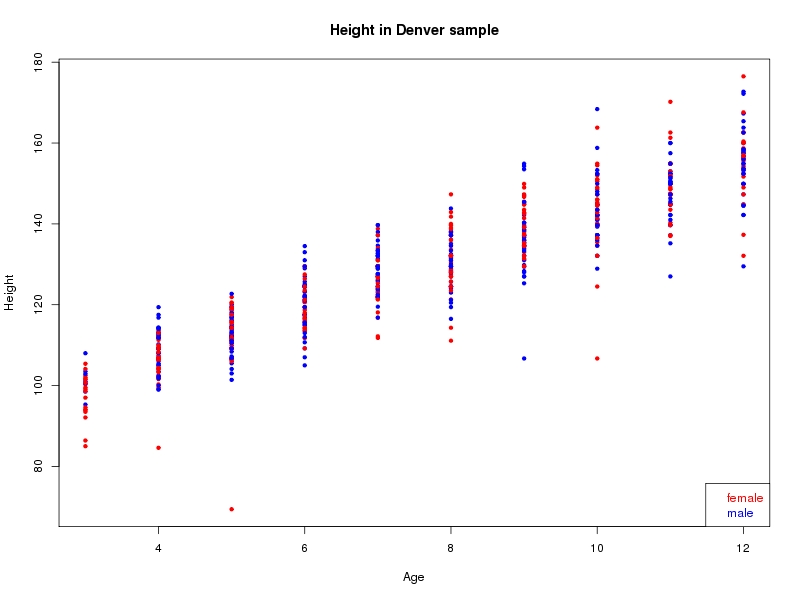
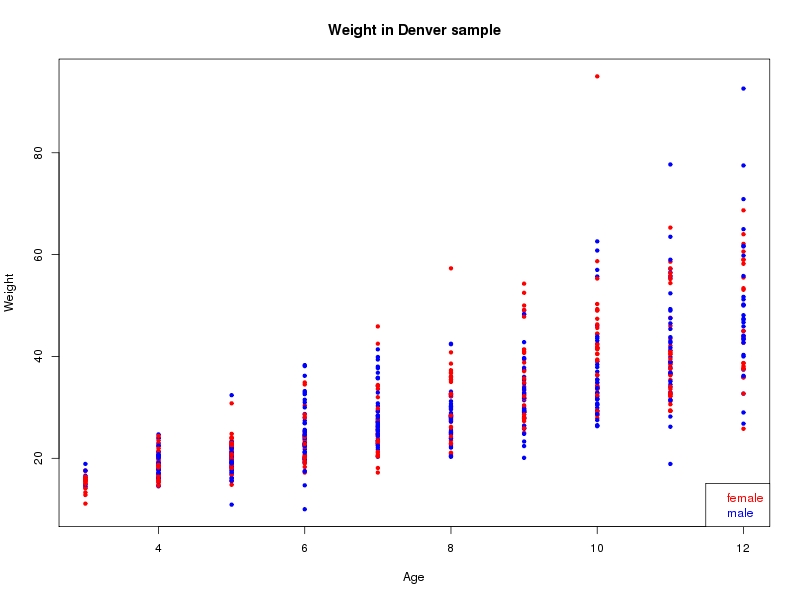
The sample for this study consisted of 3118 individuals of European ancestry belonging to two US cohorts: the “Pittsburgh” cohort (2447 participants, aged 3-49 years, largely recruited as part of the 3D Facial Norms project). The “Denver” cohort included 671 participants, aged 3-12 years, recruited as part of FaceBase project “Genetic Determinants of Orofacial Shape and Relationship to Cleft Lip/Palate” (PI: Richard A. Spritz). Each participant had his or her facial surface captured using 3D stereophotogrammetry using either the 3dMD imaging system (Pittsburgh) or Creaform Gemini system (Denver). Landmarking of facial surfaces was completed manually (Pittsburgh) or with an automated landmarking approach (https://www.facebase.org/facial_landmarking/). Additional details on recruitment and phenotyping can be found in Shaffer et al. (2016) and on the FaceBase website.
Prior to genetic association analysis, each of the 20 linear distance measures was adjusted for the effects of sex, age, age2, height, weight, and facial size (calculated as the geometric mean of the linear distance measures) using linear regression. Linear models were then used to test genetic association between each adjusted phenotype (i.e., residuals) and each SNP, under the additive genetic model, while simultaneously adjusting for the first four principal components of ancestry. Analyses were performed separately in the Pittsburgh and Denver cohorts, and combined via inverse variance-weighted meta-analysis. In order to appropriately model SNP effects, we required that the minor allele be present in at least 30 participants. This corresponds to a minor allele frequency threshold of 0.6% in the Pittsburgh cohort and 2% in the Denver cohort. SNPs meeting the minor allele count criterion in both Pittsburgh and Denver cohorts were included in the meta-analysis.
Descriptive Statistics
Please select cohort.
Pittsburgh:
Detailed 3D Facial Norms summary statistics can be found on their FaceBase Summary Page.
Explore Project Data
This project includes two independent cohorts, each comprised of unrelated individuals of self-described European white ancestry from the United States. The first cohort (the Pittsburgh cohort) was comprised of 2447 unrelated individuals ranging in age from three to 49 years. The second cohort (the Denver cohort) was comprised of 671 unrelated individuals ranging in age from three to 12 years. Each sample was analyzed individually and then combined in a meta-analysis.
All genomic coordinates are mapped to Human GRCh37 - hg19 genome assembly.
Custom Plots
Please select one option from each of the categories. You must select one option from each available category to be able to submit the request.Publications
The 3D Facial Norms Database: Part 1. A Web-Based Craniofacial Anthropometric and Image Repository for the Clinical and Research Community.
Weinberg SM, Raffensperger ZD, Kesterke MJ, Heike CL, Cunningham ML, Hecht JT, Kau CH, Murray JC, Wehby GL, Moreno LM, Marazita ML.
Cleft Palate Craniofac J. 2015 Oct 22. [Epub ahead of print]
PMID: 26492185
Using the 3D Facial Norms Database to investigate craniofacial sexual dimorphism in healthy children, adolescents, and adults.
Kesterke MJ, Raffensperger ZD, Heike CL, Cunningham ML, Hecht JT, Kau CH, Nidey NL, Moreno LM, Wehby GL, Marazita ML, Weinberg SM.
Biol Sex Differ. 2016 Apr 22;7:23. doi: 10.1186/s13293-016-0076-8. eCollection 2016.
PMID: 27110347