Human Genomics Analysis Interface
dbGaP Study Accession: phs000774.v2.p1.
Principal Investigator: Mary L. Marazita, University of Pittsburgh, Pittsburgh, PA, USA
Funding Sources:
X01-HG007485 Genetics of Orofacial Clefts and Related Phenotypes. National Human Genome Research Institute, CIDR Genotyping, National Institutes of Health, Bethesda, MD, USA
R01-DE-016148, Extending the Phenotype of Nonsyndromic Orofacial Clefts. National Institute of Dental and Craniofacial Research, National Institutes of Health, Bethesda, MD, USA
R01-DE-012472, HOMOZYGOSITY MAPPING OF ORAL-FACIAL CLEFTS IN TURKEY. National Institute of Dental and Craniofacial Research, National Institutes of Health, Bethesda, MD, USA
R01-DE-009886, GENE MAPPING STUDIES OF ORAL/FACIAL CLEFTS - CHINA/INDIA. National Institute of Dental and Craniofacial Research, National Institutes of Health, Bethesda, MD, USA
P50-DE-016215, CRANIOFACIAL ANOMALIES RESEARCH CENTER. National Institute of Dental and Craniofacial Research, National Institutes of Health, Bethesda, MD, USA
K99-DE-022378, GENETIC STUDIES OF NONSYNDROMIC CLEFTS IN POPULATIONS OF AFRICAN DESCENT. National Institute of Dental and Craniofacial Research, National Institutes of Health, Bethesda, MD, USA
R37-DE-008559, MOLECULAR GENETIC EPIDEMIOLOGY OF CLEFT LIP AND PALATE. National Institute of Dental and Craniofacial Research, National Institutes of Health, Bethesda, MD, USA
R01-DE-014667, CLEFT LIP GENETICS: A MULTI CENTER INTERNATIONAL CONSORTIUM. National Institute of Dental and Craniofacial Research, National Institutes of Health, Bethesda, MD, USA
R01-DD-000295, HEALTH OUTCOMES AND IMPROVED PHENOTYPIC CHARACTERIZATION OF CLEFT LIP AND PALATE. National Center on Birth Defects and Developmental Disabilities, Centers for Disease Control and Prevention, Atlanta, GA, USA
R21-DE-016930, PLANNING INTERNATIONAL OROFACIAL CLEFT GENETIC STUDIES. National Institute of Dental and Craniofacial Research, National Institutes of Health, Bethesda, MD, USA
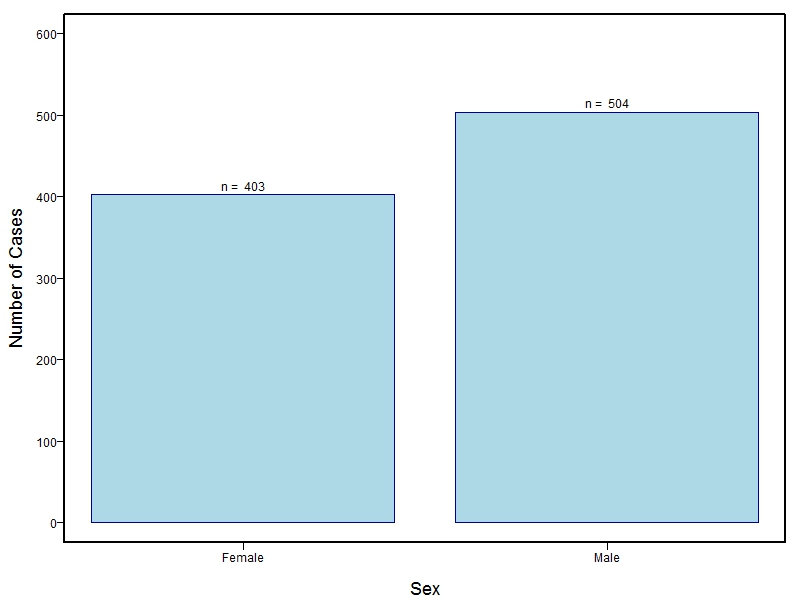
The current study is the culmination of many years of ongoing collaborations among the study investigators (for complete list of investigators, see dbGaP). Molecular genetic studies of orofacial clefts (OFC) by members of this research consortium began in the 1980's, followed by genome-wide linkage studies in the 1990's-early 2000's, and then genome-wide association studies in the 2010's. A rich phenotyping approach was first added to the consortium in 1999 as a project led by Dr. Mary Marazita under a center grant headed by Dr. Jeff Murray. Additional sites were added throughout the 2000's, until multiple populations and a large number of individuals (~12,000) comprise the current study population for genetic studies of OFCs and related subclinical phenotypes—subtle features believed to represent mild manifestations of the same underlying genetic susceptibility responsible for OFCs. As such, their inclusion in case-control and family-based genetic studies can help to clarify and refine the relationship between genotype and phenotype. Information on how to obtain individual level data is available on dbGaP.
Methods
Genotyping and quality control methods are available on dbGaP and are briefly described in Leslie et al. “A genome-wide association study of nonsyndromic cleft palate identifies an etiologic missense variant in GRHL3”. Am J Hum Genet. 2016; and Leslie et al. “A multi-ethnic genome-wide association study identifies novel loci for nonsyndromic cleft lip with or without cleft palate on 2p24.2, 17q23 and 19q31”. Hum Mol Genet. 2016.
Briefly, GWAS scans were conducted in two subsets: independent cases and controls, and an independent set of case-parent trios. Statistical analysis was performed in PLINK using logistic regression while adjusting for principal components of ancestry and the transmission disequilibrium test. Association results for the two GWAS scans were combined by weighted odds ratio meta-analysis to estimate study-wide association results. Analyses were completed in the full multiethnic sample or stratified by genetically defined subpopulations.
Explore Project Data
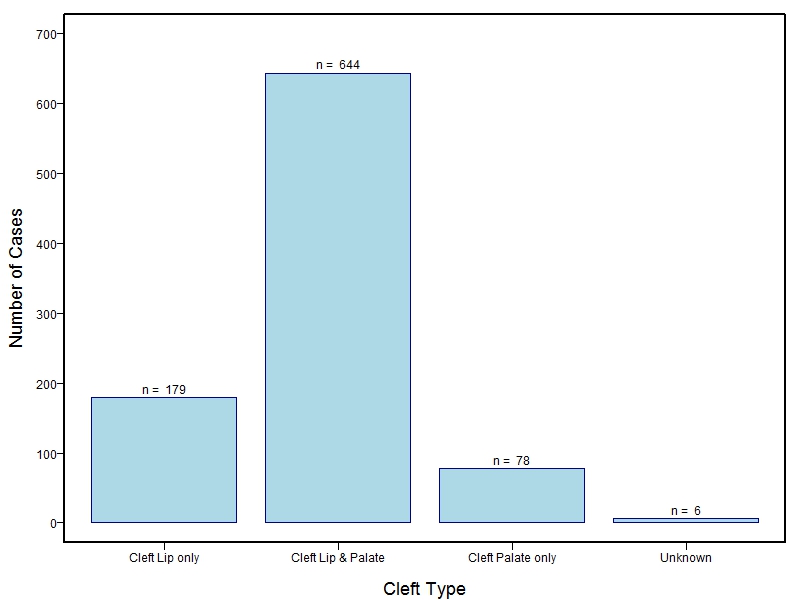
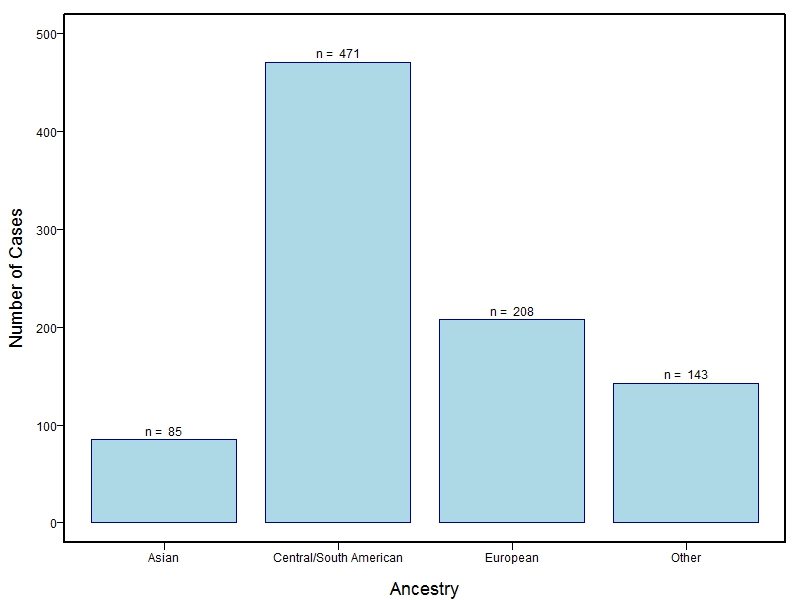
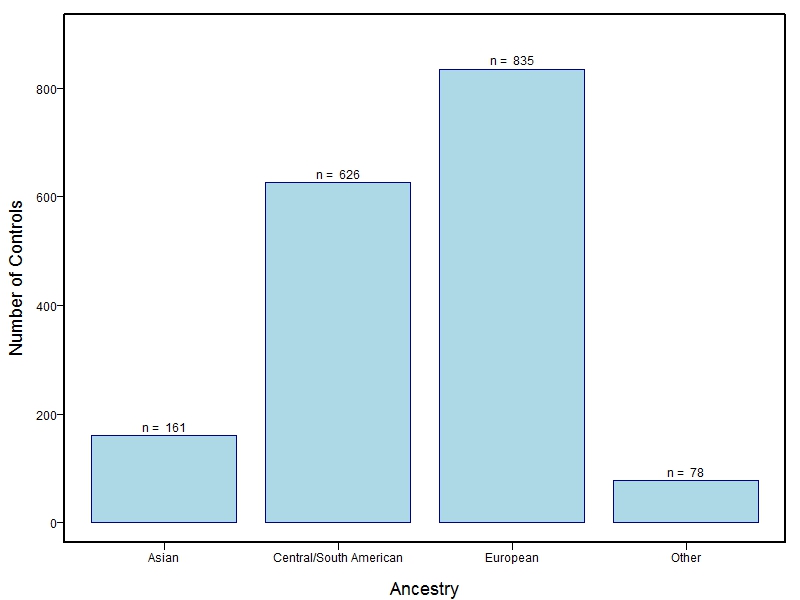
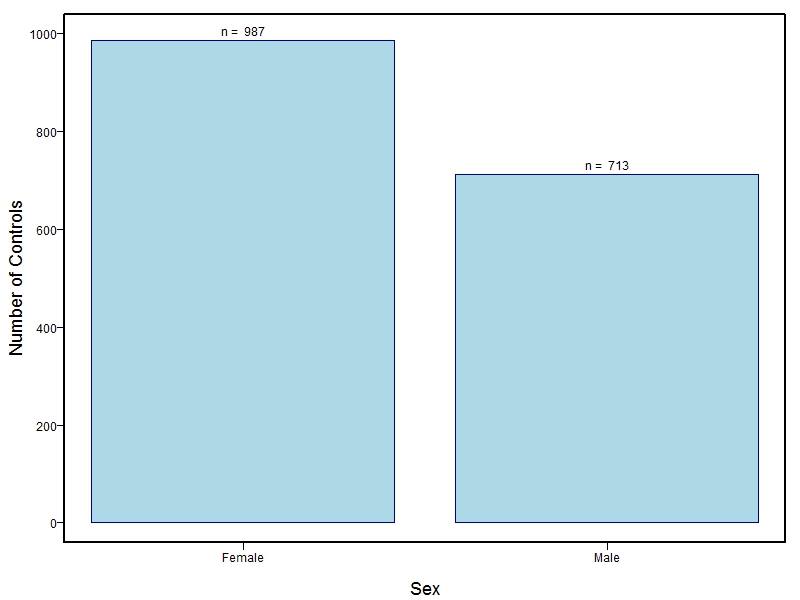
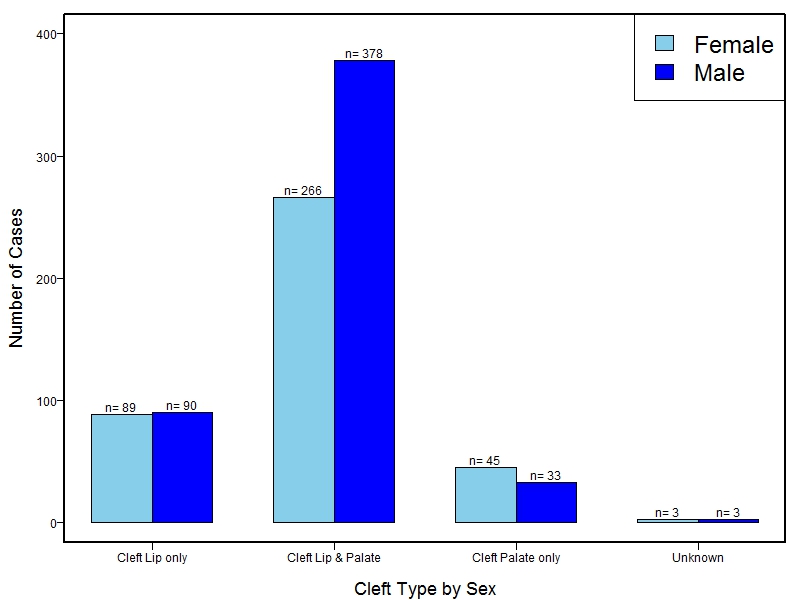
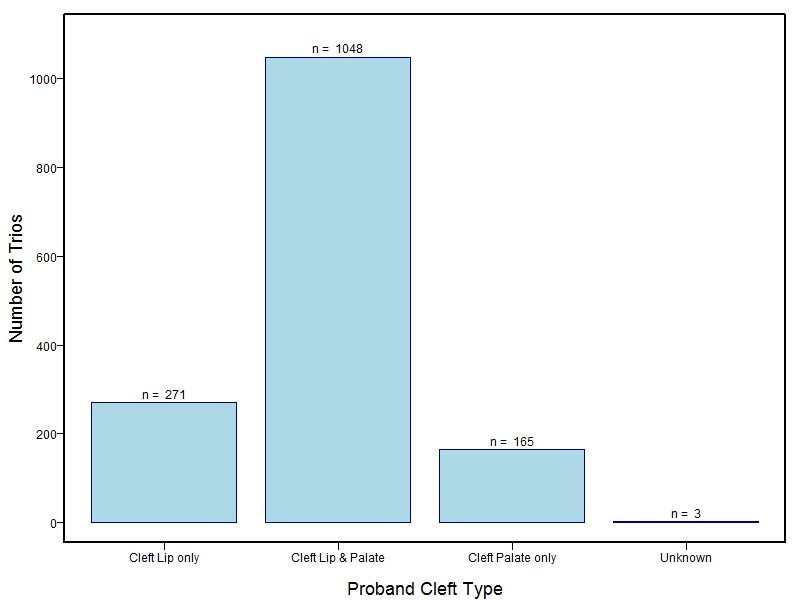
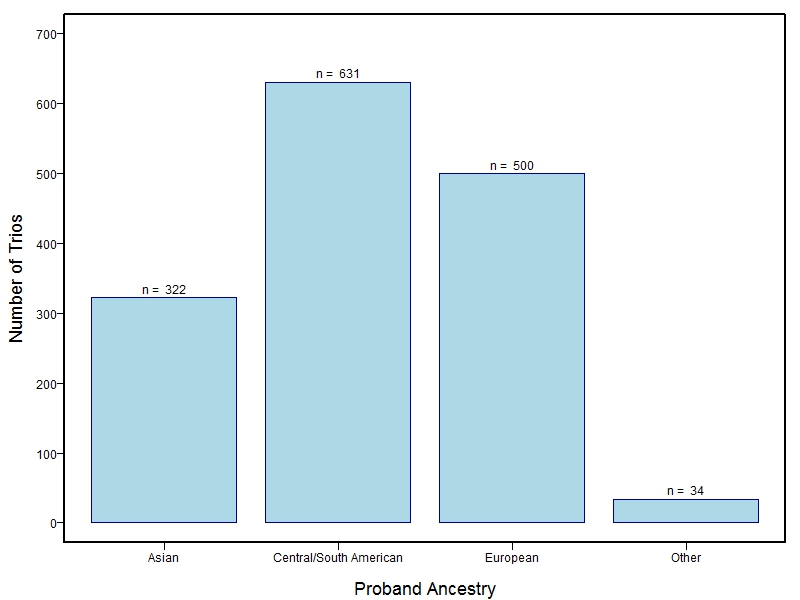
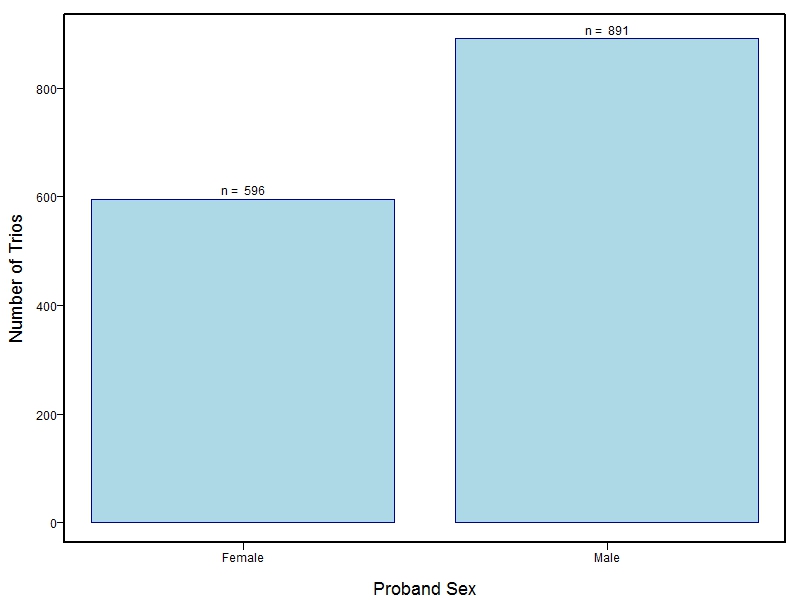
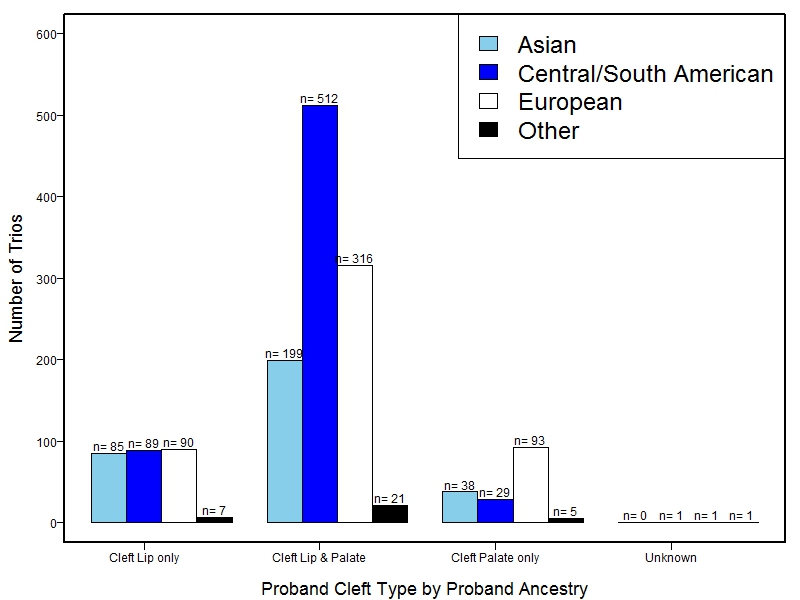
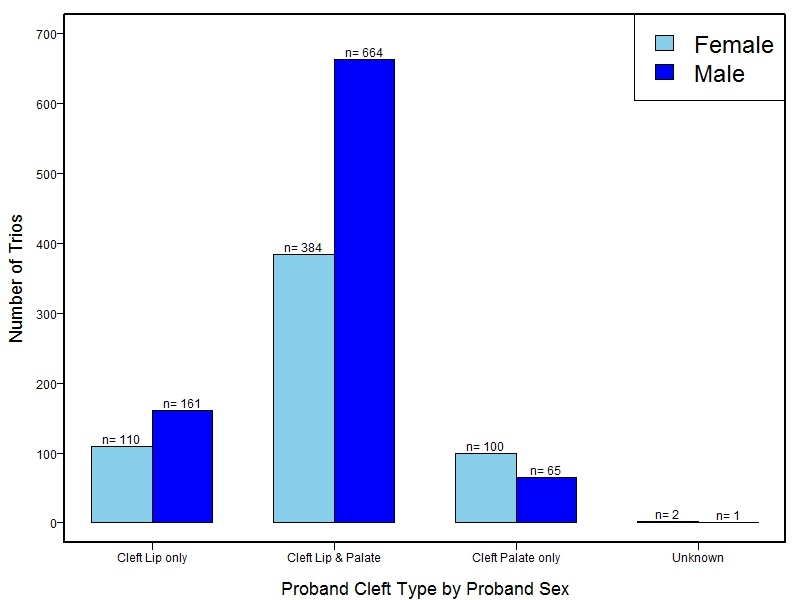
The samples for this study come from three primary ancestry groups: Europeans (from the United States, Denmark, Hungary, Spain, and Turkey), Asians (from China, the Philippines, and India), Central/South Americans (from Puerto Rico, Guatemala, Colombia, and Argentina), and Africans (from Nigeria and Ethiopia). The subjects have been assessed for various cleft types and related subclinical phenotypes. Our results database contains the GWAS results from this study of OFCs and can be explored as one study or subset into separate groups.
All genomic coordinates are mapped to Human GRCh37 - hg19 genome assembly.
Custom Plots
Publications
PubMed list for all publications associated with this project.