Human Genomics Analysis Interface
dbGaP Study Accession: phs000625.v1.p1
Principal Investigators:
Jeffrey C. Murray, University of Iowa, Iowa City, IA, USA
Mary L. Marazita, University of Pittsburgh, Pittsburgh, PA, USA
Richard K. Wilson, Washington University School of Medicine, St. Louis, MO, USA
George Weinstock, Jackson Laboratory for Genomic Medicine, Farmington, CT, USA
Funding Source: U01-HG005925. Sequencing of Significant Signals from Cleft Lip GWAS. National Institutes of Health, Bethesda, MD, USA.
Nonsyndromic cleft lip and palate (NSCL/P) is a complex disorder caused by both genetic and environmental factors and has been the focus of an extensive effort to identify genetic risk factors. A number of candidate gene studies have been performed but have not been widely replicated. To date, four independent genome wide association studies have been performed as well as a meta-analysis. Together these studies have identified many loci associated with NSCL/P. The goal of this project is to use targeted sequencing to further characterize these regions and to progress from the association signals identified by GWAS to the identification of causative genes and/or variants.
This study is part of the GWASeq project, a collaboration of five disease studies, which will sequence genomic regions from GWAS to characterize the genetic variation underlying these diseases and to compare study design and methods for the follow-up of GWAS studies by sequencing.
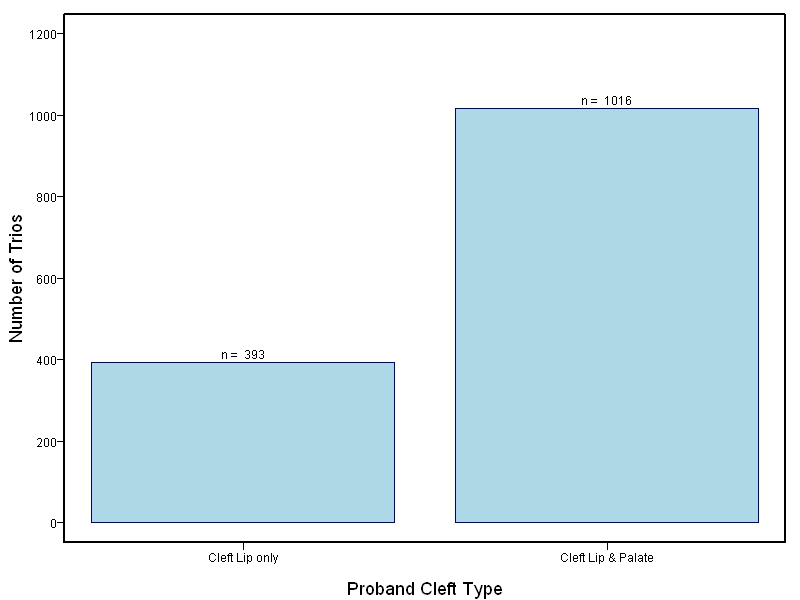
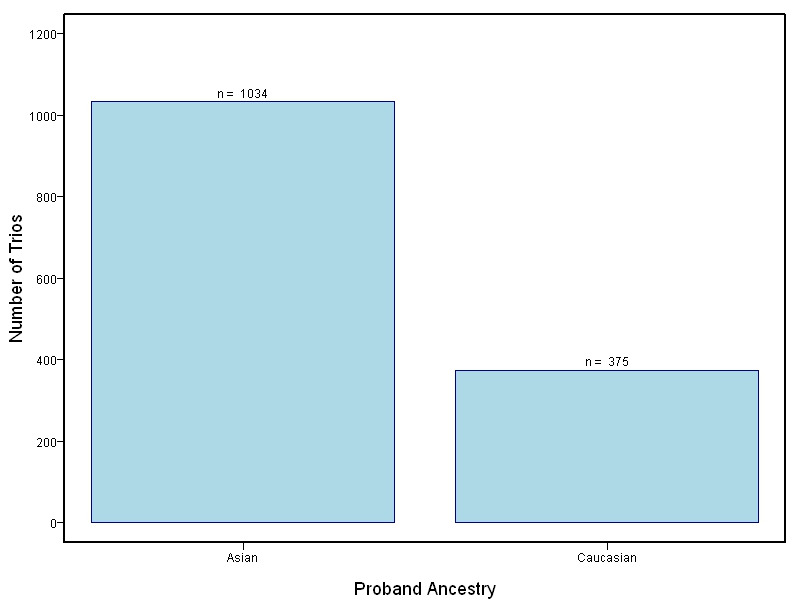
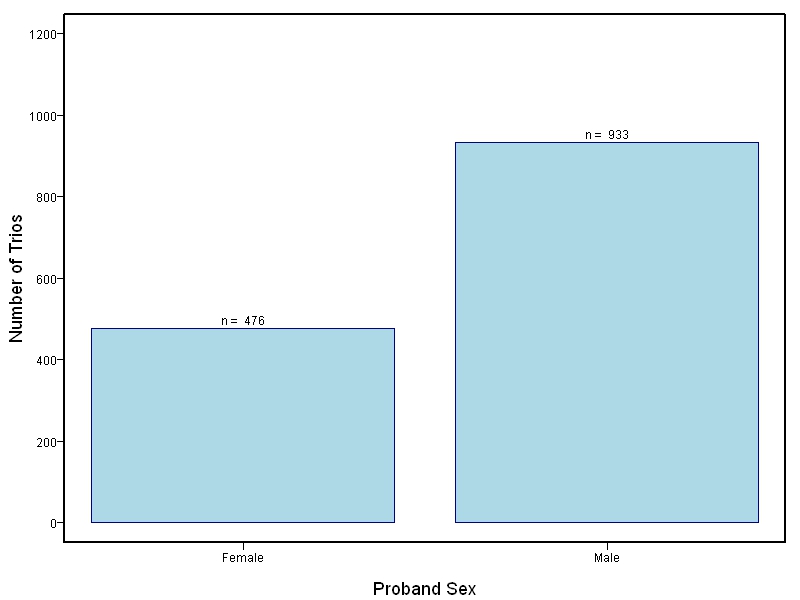
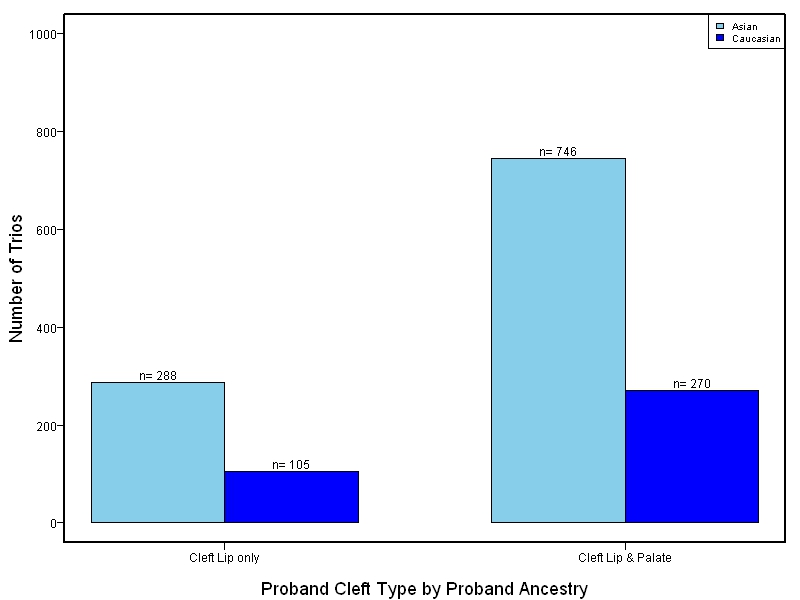
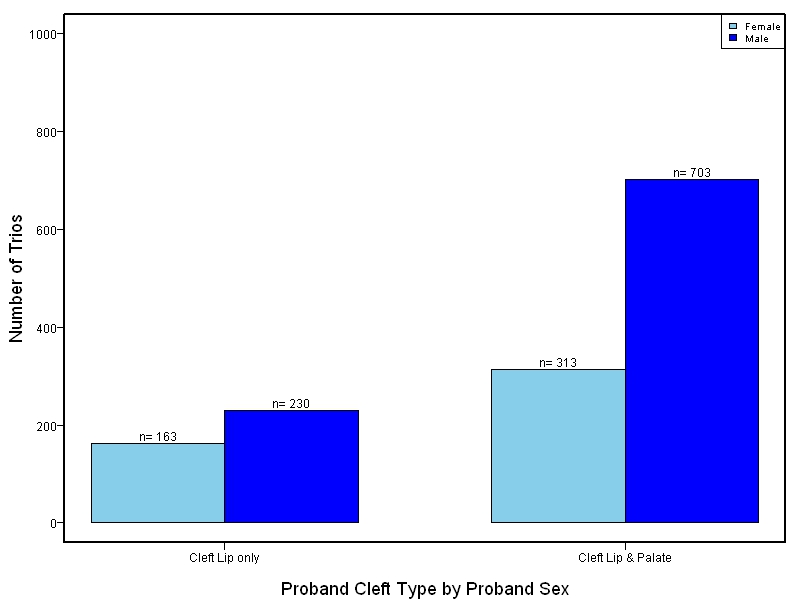
The goal of this study is to sequence 1000+ NSCL/P case-parent trios from China and the Philippines and 400 trios of European ancestry. Targeted sequencing was performed on intervals ranging between 60kb to 1Mb surrounding 13 genes/loci previously associated with NSCL/P including: IRF6, MAFB, ARHGAP29, 8q24, PAX7, VAX1, NTN1, NOG, FOXE1, MSX1, BMP4, FGFR2, PTCH1.
Sequencing was performed at the Genome Institute at Washington University.
Methods
Sequencing and quality control methods were described in Leslie et al. Identification of functional variants for cleft lip with or without cleft palate in or near PAX7, FGFR2, and NOG by targeted sequencing of GWAS loci. Am J Hum Genet. 2015 Mar 5;96(3):397-411.
Transmission disequilibrium tests (TDT) were performed in PLINK (v1.07). Inclusion criteria for SNPs were minor allele frequencies greater than 1%, genotyping efficiency greater than 95%, and Hardy-Weinberg Equilibrium p-values greater than 0.0001.
Regional association plots were created using LocusZoom. Linkage disequilibrium between the top SNP in the region and the other SNPs was determined using 1000 Genomes with either Asians (ASN) or European (EUR) reference panels, as appropriate.
Genotypic methodology is available at dbGap.
Explore Project Data
This project involved targeted sequencing of GWAS loci for CL/P. Thirteen regions were sequenced. The samples for this study come from two primary ancestry groups: Asians (from China and the Philippines) and Europeans (from several sites in the United States. Our results database contains the association results for each of the thirteen sequenced regions and can be explored in the combined sample or in each ancestry group separately. Only small targeted regions are shown.
All genomic coordinates are mapped to Human GRCh37 - hg19 genome assembly.