Human Genomics Analysis Interface
dbGaP Study Accession: phs000094.v1.p1.
Principal Investigator: Terri H. Beaty, Johns Hopkins School of Public Health, Baltimore, MD, USA
Funding Source: U01-DE018993. International Consortium to Identify Genes and Interactions Controlling Oral Clefts. National Institutes of Health, Bethesda, MD, USA
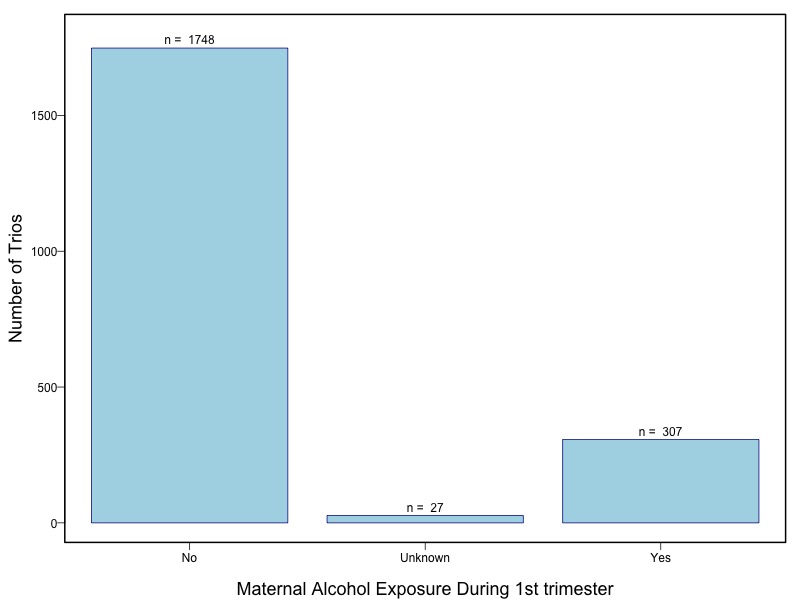
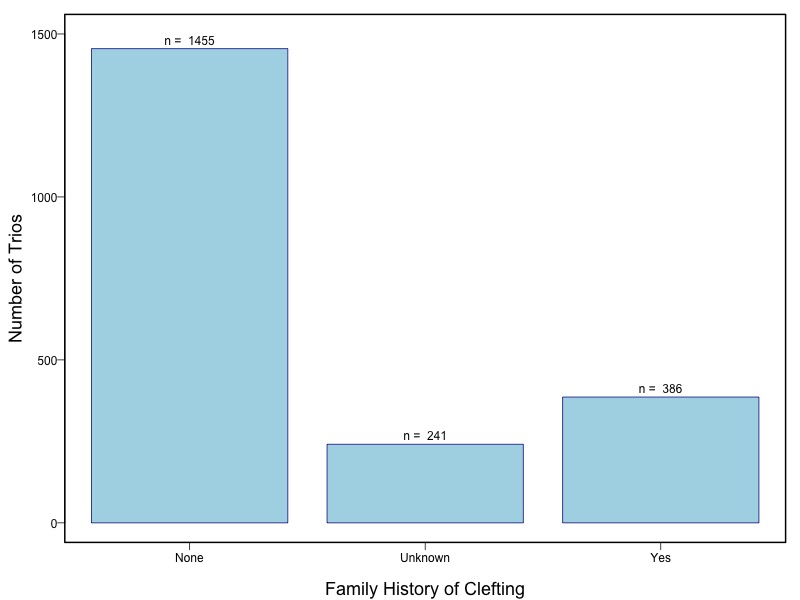
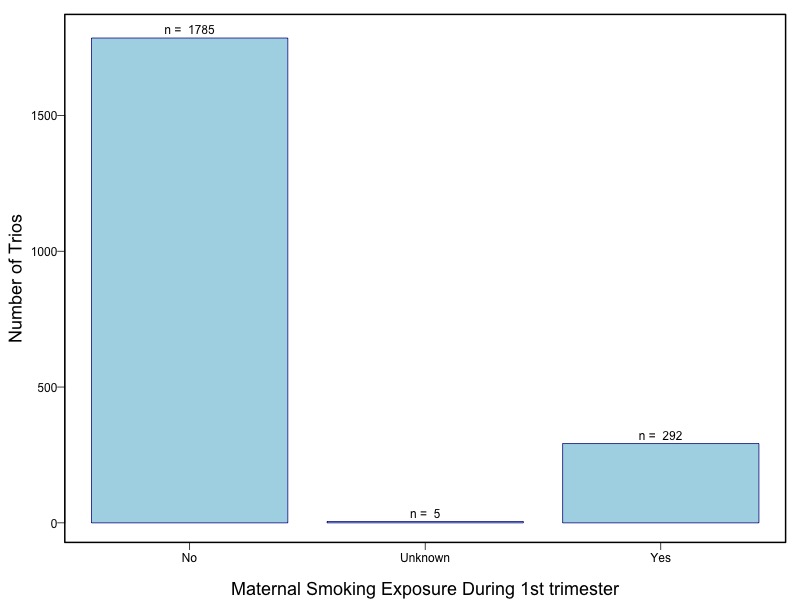
Oral clefts represent the most common group of craniofacial birth defects in humans, and include cleft lip with or without cleft palate (CL/P) and cleft palate (CP). Oral clefts have a complex and heterogeneous etiology, with strong evidence for both genetic and environmental causal factors. Candidate gene studies and genome wide linkage studies have yielded compelling but inconsistent evidence that multiple genes control risk, and several studies have shown evidence for interaction between genes and environmental exposures, especially maternal smoking and nutrient intake. This consortium assembled a large collection of cases and their parents from multiple populations for genome wide association studies as part of the Gene Environment Association Studies initiative (GENEVA), which was developed through the trans-NIH Genes, Environment, and Health Initiative (GEI). Genotyping was performed at the Johns Hopkins University Center for Inherited Disease Research (CIDR). The study was supported by the National Institute of Dental and Craniofacial Research (NIDCR). Data cleaning and harmonization were done at the GEI-funded GENEVA Coordinating Center at the University of Washington.
Information on how to obtain individual level data is available on dbGap.
Methods
Phenotypic and genotypic methodology for this study can be found on dbGap.
Transmission Disequilibrium Tests (TDT) were performed in PLINK (v1.07)1. Inclusion criteria for SNPs were minor allele frequencies greater than 5% and Hardy-Weinberg Equilibrium p-values greater than 0.0001.
Manhattan plots were created using the qqman2 package in R (v.3.x).
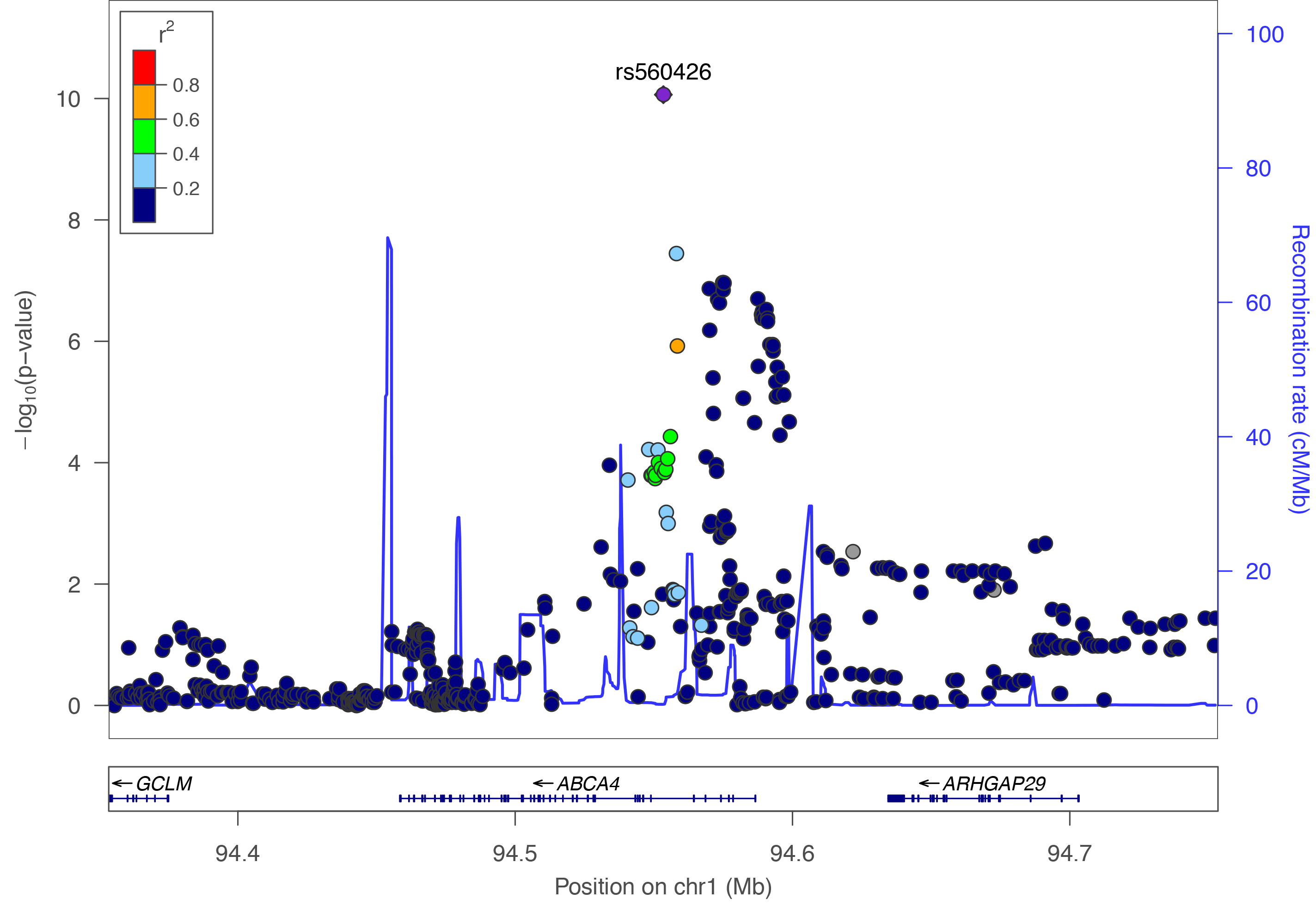
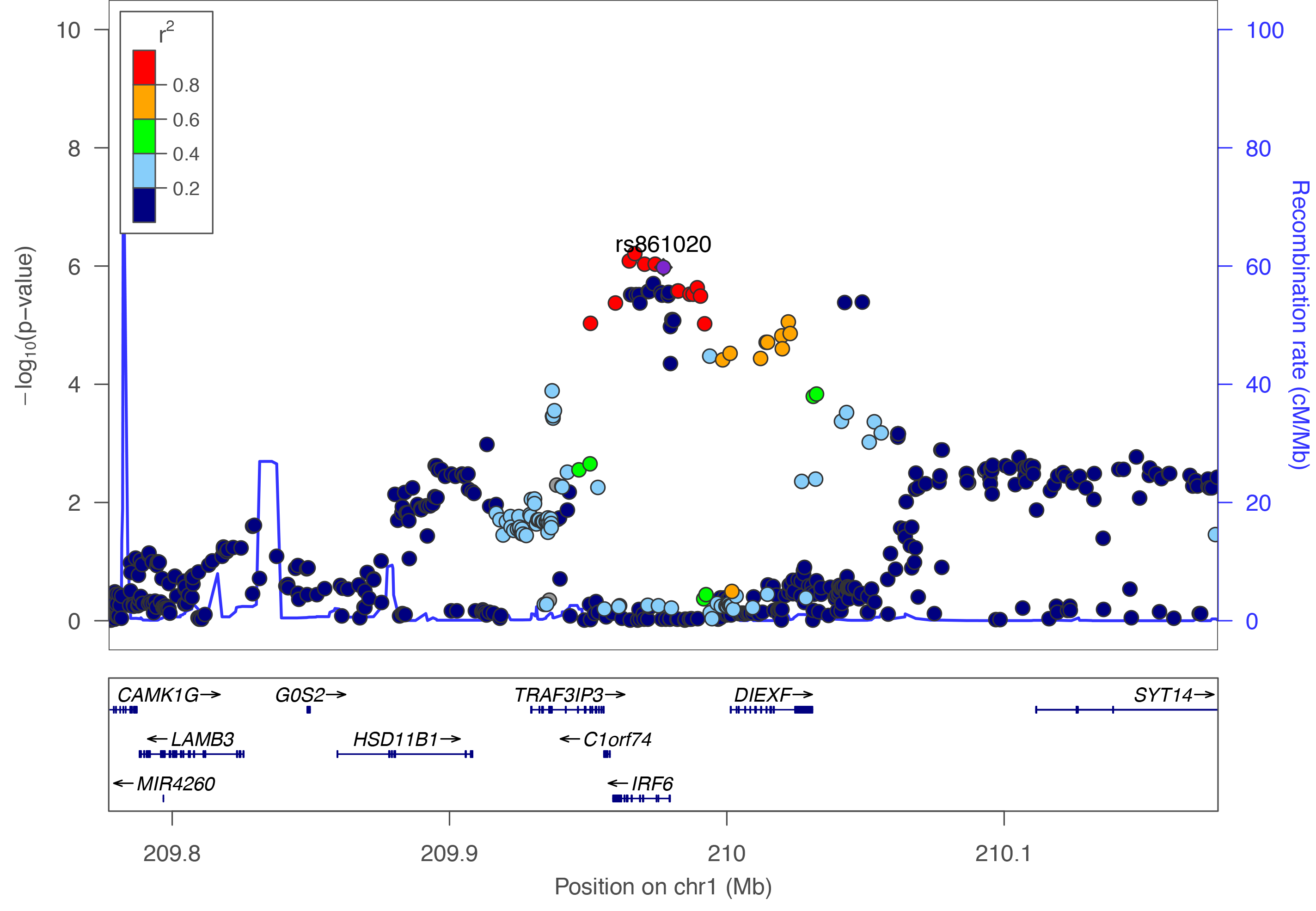
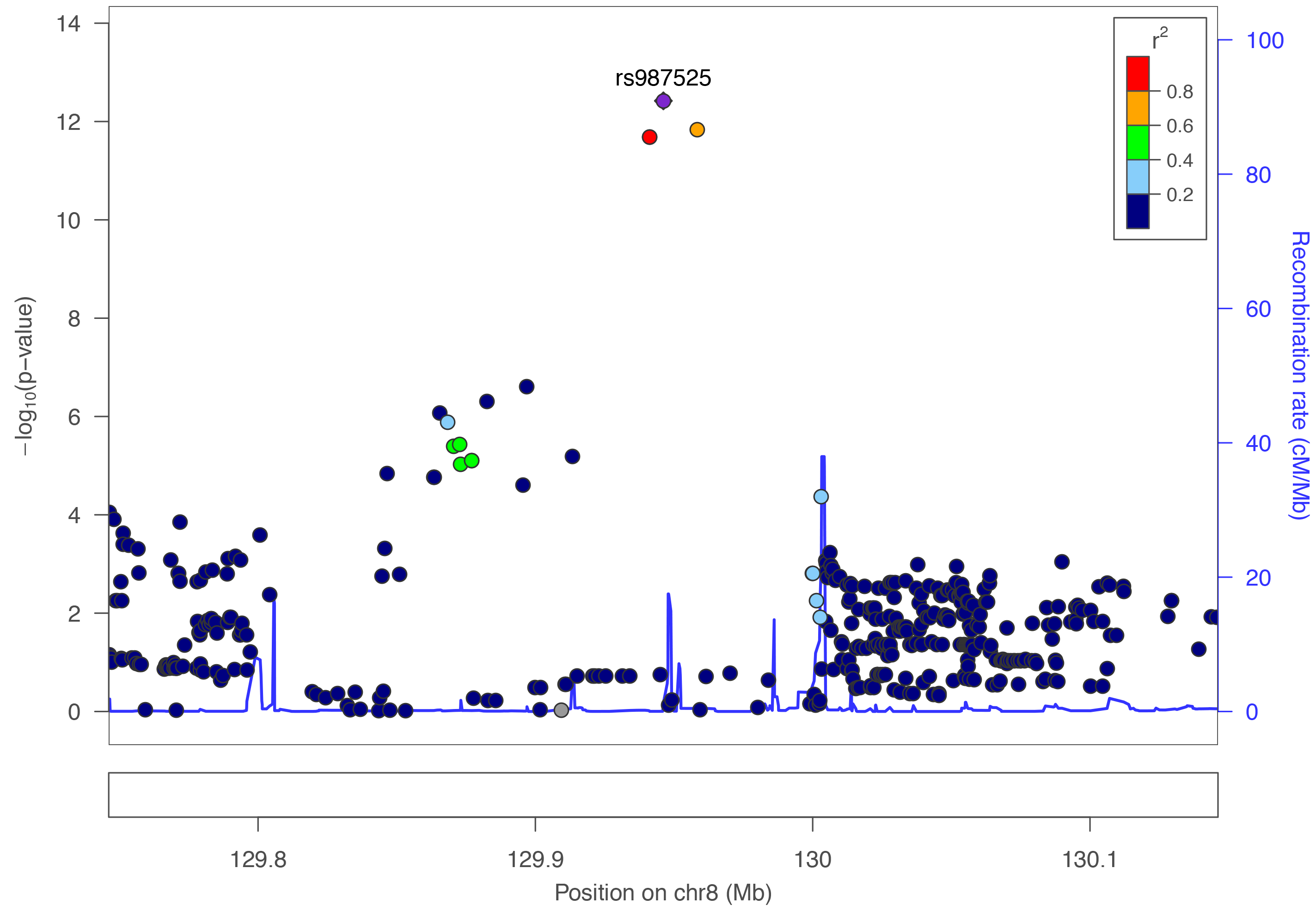
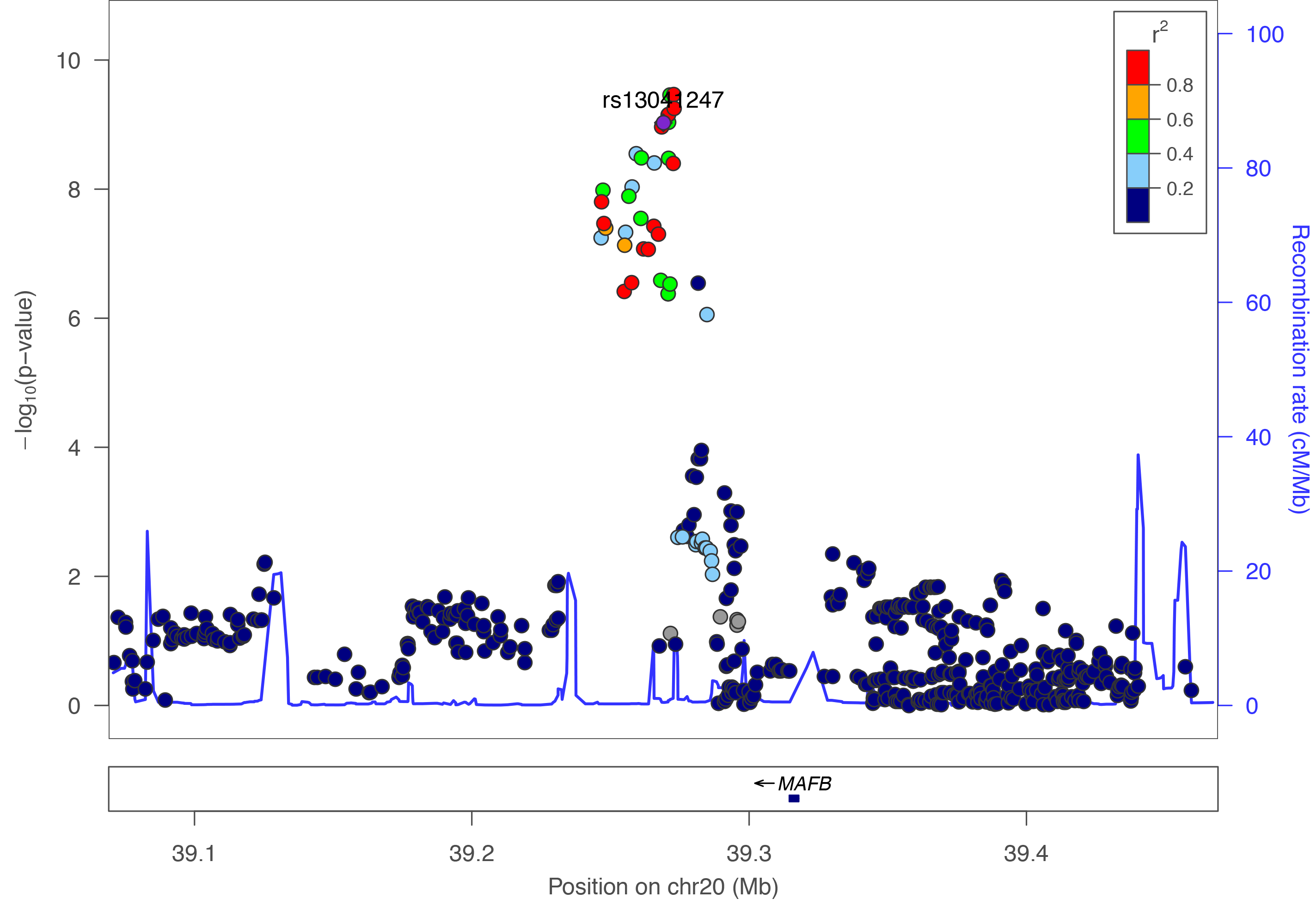
Regional plots created using LocusZoom.
1 Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ & Sham PC (2007). PLINK: a toolset for whole-genome association and population-based linkage analysis. American Journal of Human Genetics, 81.
2 Turner, S.D. qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. biorXiv DOI: 10.1101/005165 (2014).
Explore Project Data
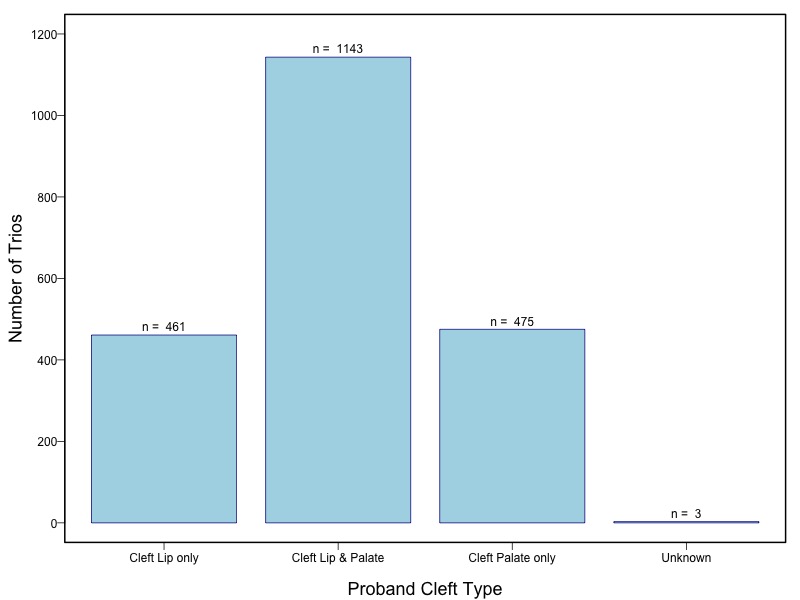
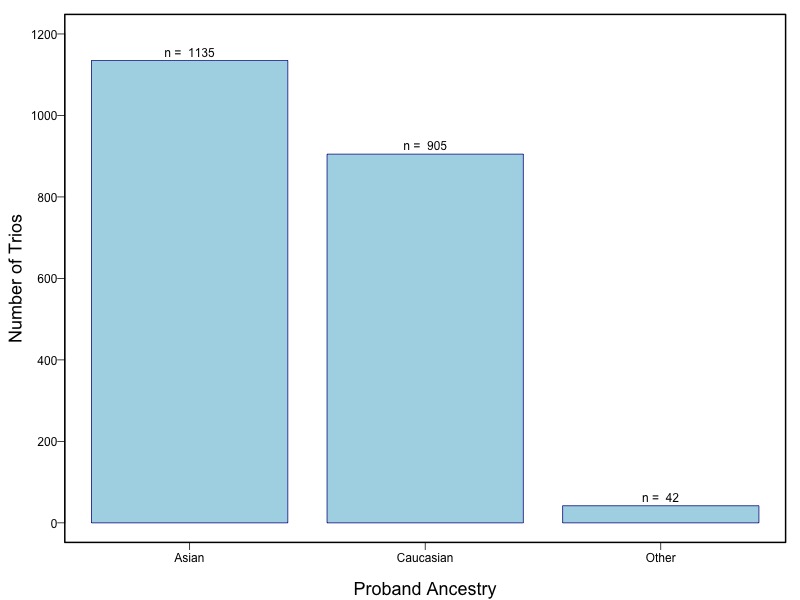
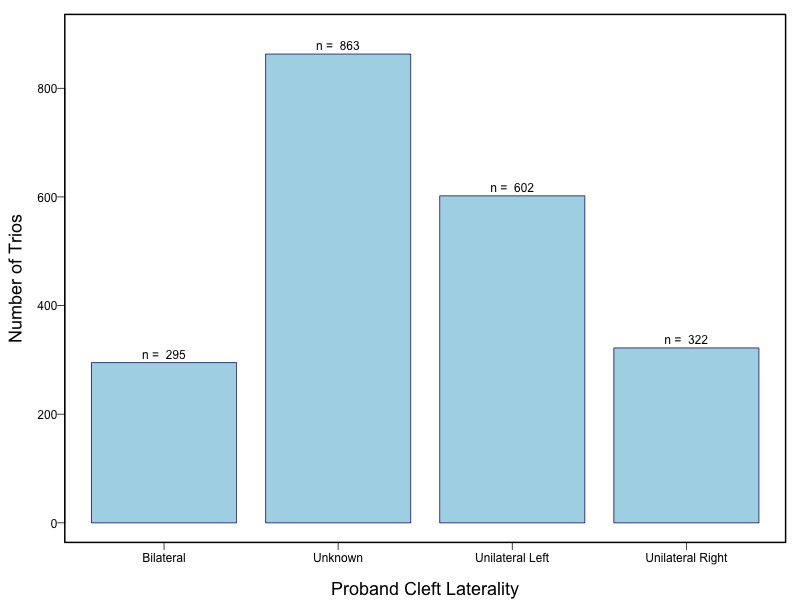
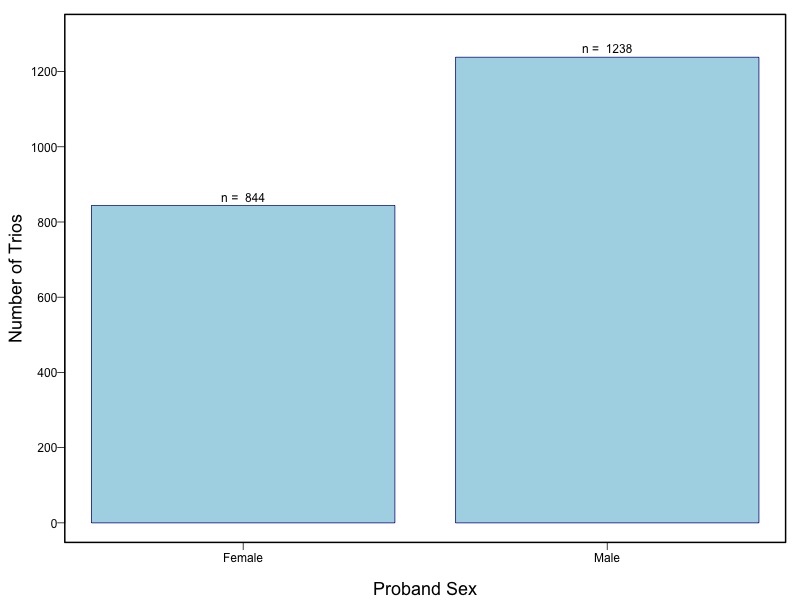
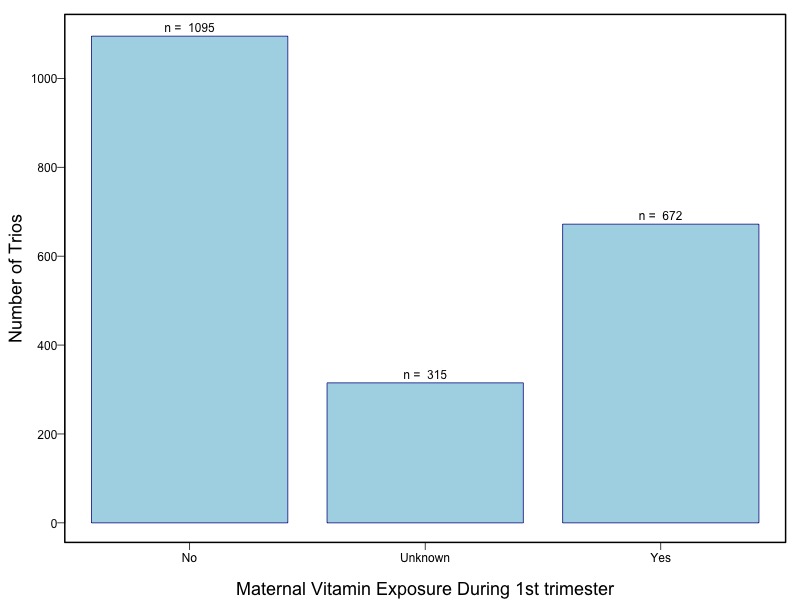
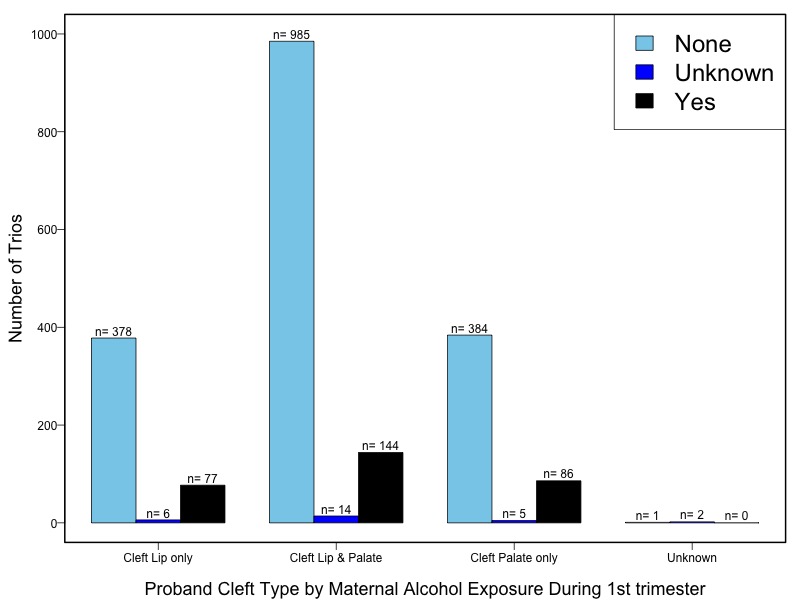
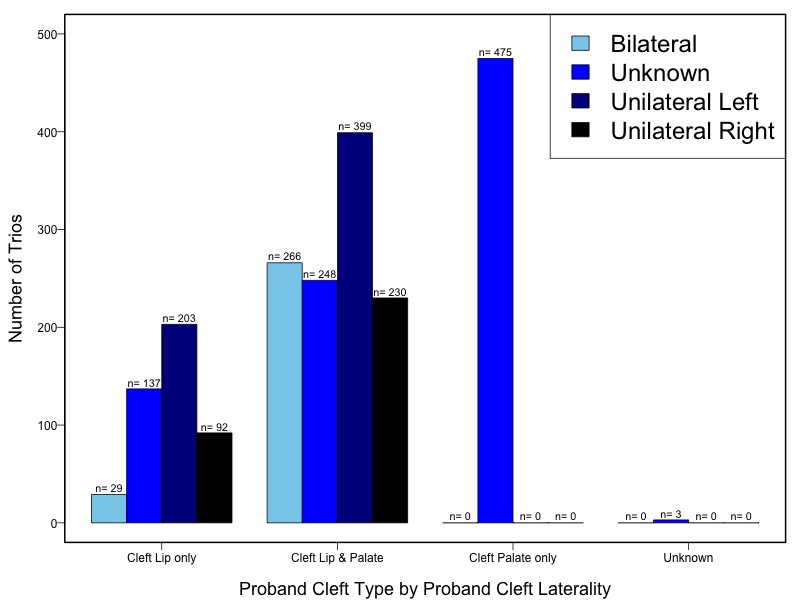
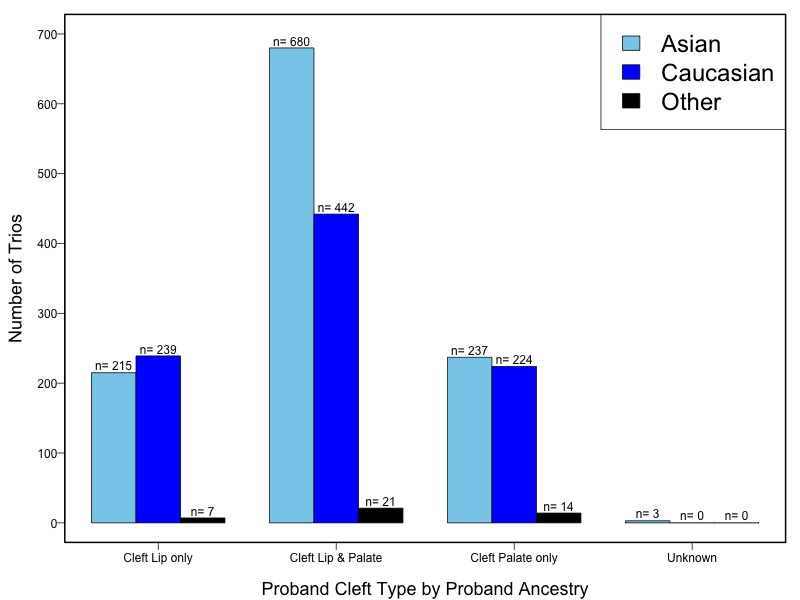
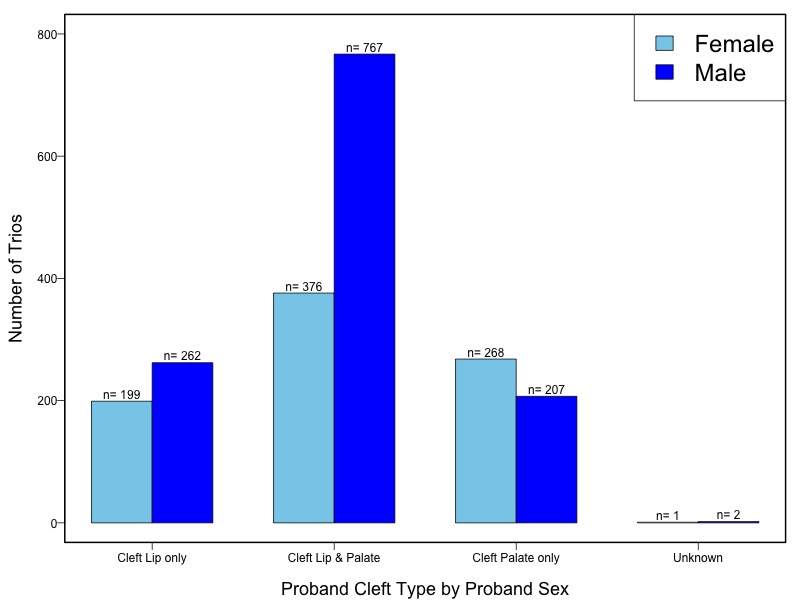
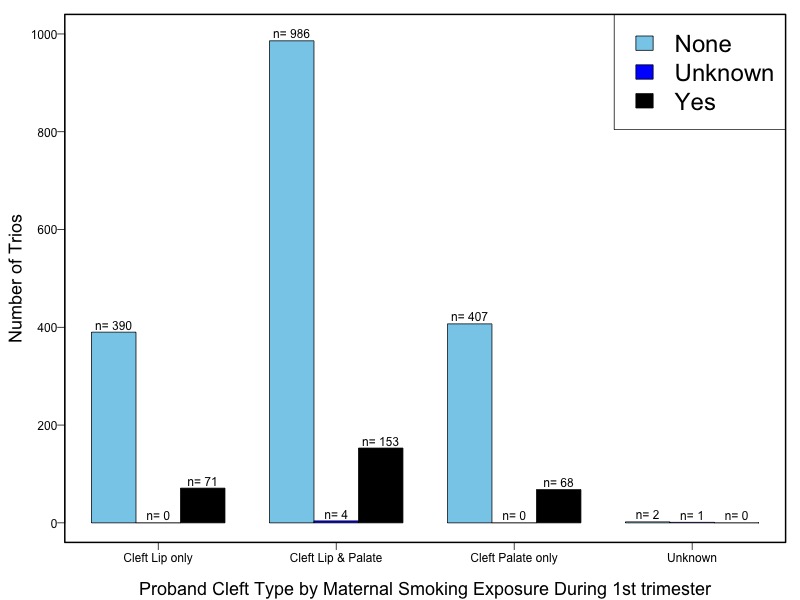
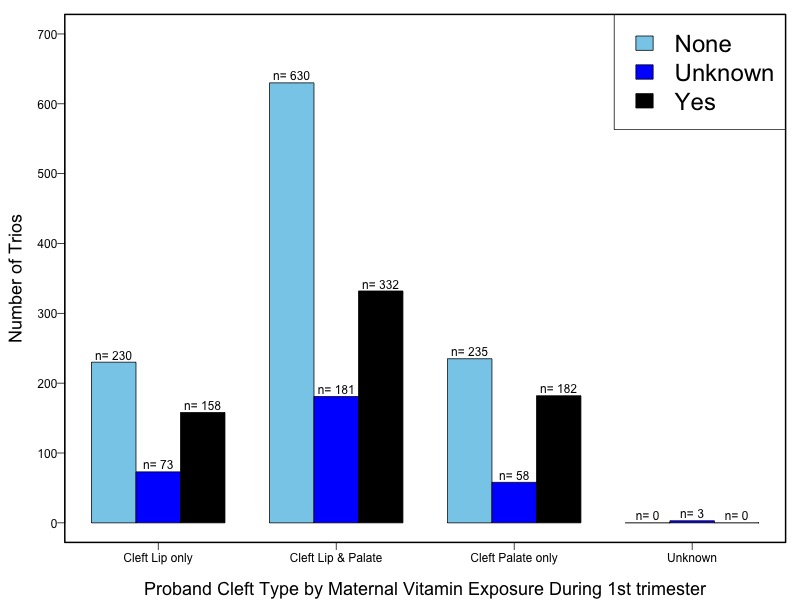
The samples for this study come from two primary ancestry groups: Asians (from China, the Philippines, Singapore, Korea, and Taiwan) and Europeans (from the United States, Norway, and Denmark). The subjects have been assessed for various cleft types and reported on first trimester environmental exposures. Our results database contains the GWAS results from this study of CL/P and can be explored as one study or subsetted into separate groups.
All genomic coordinates are mapped to Human GRCh37 - hg19 genome assembly.
View published peaks
These are peaks that have been published in Nature Genetics. They are for all ancestries, all cleft types, and for imputed data.Custom Plots
Please select one option from each of the categories. You must select one option from each available category to be able to submit the request.Publications
PubMed list for all publications associated with grant number U01-DE018993.